Abstract
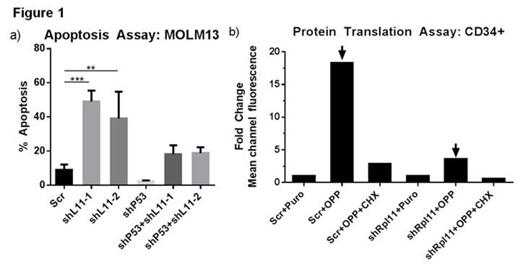
Therapy-related myelodysplastic syndrome (t-MDS) is a lethal complication of cytotoxic cancer treatment and a leading cause of non-relapse mortality among patients undergoing autologous hematopoietic cell transplantation (aHCT) for lymphoma. The molecular abnormalities leading to t-MDS, however, are poorly understood. We are studying a prospective, longitudinal cohort of patients undergoing aHCT for lymphoma at our center with serial sample collection and banking. Using this approach we have identified impaired hematopoietic function and altered gene expression in CD34+ progenitor cells from pre-aHCT samples from patients who later developed t-MDS (cases) as compared to patients who did not develop t-MDS (controls) (Cancer Cell 2011, 20:591). These results indicate that genetic programs associated with t-MDS are perturbed long before disease onset. The alterations seen include down-regulation of genes related to ribosome biogenesis and translation, highlighting the potential contribution of altered ribosome function to disease development. Interestingly, a group of inherited ribosomal disorders including Diamond-Blackfan Anemia (DBA), called ribosomopathies, are characterized by cytopenias and a propensity for MDS development, features that closely resemble those of t-MDS patients. We evaluated the expression level of 79 ribosomal genes and a selected group of ribosome associated genes in cases and controls by multiplex Q-RT-PCR. Expression of DBA associated genes RPL10, RPL11, and RPL27 was significantly reduced in cases compared to controls. Each of these genes is reported to be associated with DBA. In addition, expression of p53, p21, c-Myc, and hdm2 were significantly upregulated in t-MDS cases. Altered expression of RPL11 is hypothesized to contribute to hematopoietic dysfunction by modulating the p53 ubiquitin ligase, HDM2, and activating p53-dependent apoptosis. We therefore characterized the effect of reduced RPL11 expression on hematopoietic function and determined the role of p53 activation in mediating the effects of RPL11 knockdown. We generated lentiviral vectors expressing shRNAs against RPL11 to reduce its expression in hematopoietic cell lines and CD34+ stem cell populations. Cells were cultured for 48 to 96 hours post-transduction followed by apoptosis analysis using Annexin-V staining, and proliferation by dye dilution assays. We confirmed >90% knockdown of RPL11 in shRNA expressing cells (MOLM13) by Q-RT-PCR and Western blot analysis. RPL11 knockdown resulted in 5-fold increase in apoptosis (Figure 1a) and 2-fold inhibition of proliferation of MOLM13 hematopoietic cells that express wild type p53, but did not affect survival and proliferation of p53 mutated HL60 cells. This suggests that cellular responses to RPL11 deficiency may depend at least in part on intact p53 signaling. This was confirmed in experiments showing that concomitant shRNA mediated knockdown of p53 and RPL11 (Figure 1a) resulted in a 2-fold reduction of apoptosis compared to RPL11 knockdown alone. Furthermore, RPL11 knockdown in CD34+ stem/progenitor cells resulted in 40% increase in apoptosis, 30% inhibition of proliferation and 90% decrease in colony forming cell (CFC) frequency in methylcellulose progenitor assays. We measured protein translation by measuring the intensity of an alkyne analog of puromycin (OPP) integrated into newly translated proteins, ligation to an azide dye, and flow cytometry analysis. We observed a 5-fold drop in global protein translation with RPL11 knockdown using the Click-iT Plus Alexa Fluor Protein Synthesis Assay (Figure 1b). Our results suggest that ribosomal abnormalities occur early in the course of t-MDS development and may lead to enhanced cell death and impaired proliferation. These results support a potential role of aberrant RPL11 expression in altered cell survival, proliferation, and differentiation, which may contribute to hematopoietic dysfunction seen early in the course of t-MDS development.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal