Abstract
Background
MicroRNAs (miRNAs) are considered to play a key role in the pathogenesis of myelodysplastic syndromes (MDS). However, the effect of miRNA and targeted mRNA on signal transduction is not fully understood in MDS.
Objective
The objective of this study is to identify the miRNAs-regulated pathways.
Methods
Affymetrix GeneChip microRNA and PrimeView Array were used to analyze miRNAs and gene expression profile of CD34+ cells in 12 MDS patients and 6 healthy controls. Comprehensive bioinformatics analysis of the coordinate expression of miRNAs and mRNAs including Difference, Go, Pathway, Pathway-network, miRNA-Gene-Network and miRNA-Go-Network analysis was performed to identify the miRNAs-regulated networks.
Results
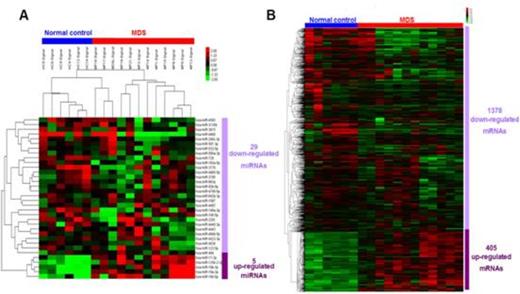
1. 34 differentially expressed miRNAs (5 up- and 29 down-regulated miRNAs) and 1783 mRNAs (405 up- and 1378 down-regulated mRNAs) in CD34+ cells from MDS and Healthy controls were identified by miRNA and mRNA microarray, respectively (Fig.1).
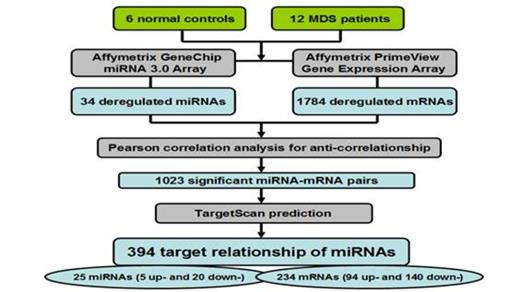
2. 25 dysregulated miRNAs and 234 targeted mRNAs were identified by a combination of Pearson's correlation analysis and prediction by TargetScan; 394 target relationship of miRNAs was established (Fig.2).
3. Go analysis revealed that these miRNA-mRNAs pairs were involved in signal transduction, apoptotic process, DNA-dependent transcription regulation, protein phosphophoration, etc. Pathway analysis showed that MAPK, JAK/STAT and PI3K/Akt signaling pathways might be regulated by these miRNA-mRNAs pairs (Fig.3).
4. The pathway-network analysis revealed that MAPK signaling pathway, Jak-Stat signaling pathway and apoptosis signaling pathway (displayed by red cycle) located in the downstream of signal networks (Fig. 3E). Dysregulation of These pathways may be more meaningful for explaining the pathogenesis of MDS.
5. Through a combination of Pathway, miRNA-Gene-Network and miRNA-Go- Network analysis, 29 miRNA-mRNA-regulated pathways were identified such as miR-148a/TEK/PI3K-Akt signaling pathway, miR-195/BDNF/MAPK signaling pathway, miR-195/DLL1/Notch signaling pathway, miR-145/CCND2/ JAK-STAT signaling pathway, etc. (Table 1).
Conclusion
Alteration expression of several miRNAs and targeted mRNAs might have an important impact on cancer-related cellular pathways including MAPK, PI3K/Akt, JAK/STAT, etc. The role of these miRNAs-mediated pathways in pathogenesis of MDS merit further investigation.
Significant miRNA-mRNA pairs identified through a integration of mcroRNA-mRNA microarray
Significant miRNA-mRNA pairs identified through a integration of mcroRNA-mRNA microarray
Parts of dysregulated miRNAs, genes and targeted pathway in MDS
| MicroRNA . | Style . | Gene_synbol . | Pathway . |
|---|---|---|---|
| miR-148a | Down | TEK | PI3K-Akt signaling pathway |
| ITGA9 | PI3K-Akt signaling pathway | ||
| KIT | PI3K-Akt signaling pathway | ||
| HMGA2 | Transcriptional misregulation in cancer | ||
| miR-145 | Down | HHEX | Transcriptional misregulation in cancer |
| MEIS1 | Transcriptional misregulation in cancer | ||
| miR-200c | Down | EFNA1 | PI3K-Akt signaling pathway |
| KLF3 | Transcriptional misregulation in cancer | ||
| miR-195 | Up | BDNF | MAPK signaling pathway |
| CDC25B | MAPK signaling pathway | ||
| DLL1 | Notch signaling pathway | ||
| MRAS | MAPK signaling pathway | ||
| miR-17 | Up | CAMK2D | Calcium signaling pathway |
| miR-19a | Up | MAML1 | Notch signaling pathway |
| SLC8A1 | Calcium signaling pathway | ||
| THBS1 | Proteoglycans in cancer | ||
| TNF | MAPK signaling pathway | ||
| TNFRSF1B | Adipocytokine signaling pathway | ||
| ACSL1 | Adipocytokine signaling pathway | ||
| EDNRB | Calcium signaling pathway | ||
| miR-19b | Up | CALM1 | Calcium signaling pathway |
| TNF | Proteoglycans in cancer |
| MicroRNA . | Style . | Gene_synbol . | Pathway . |
|---|---|---|---|
| miR-148a | Down | TEK | PI3K-Akt signaling pathway |
| ITGA9 | PI3K-Akt signaling pathway | ||
| KIT | PI3K-Akt signaling pathway | ||
| HMGA2 | Transcriptional misregulation in cancer | ||
| miR-145 | Down | HHEX | Transcriptional misregulation in cancer |
| MEIS1 | Transcriptional misregulation in cancer | ||
| miR-200c | Down | EFNA1 | PI3K-Akt signaling pathway |
| KLF3 | Transcriptional misregulation in cancer | ||
| miR-195 | Up | BDNF | MAPK signaling pathway |
| CDC25B | MAPK signaling pathway | ||
| DLL1 | Notch signaling pathway | ||
| MRAS | MAPK signaling pathway | ||
| miR-17 | Up | CAMK2D | Calcium signaling pathway |
| miR-19a | Up | MAML1 | Notch signaling pathway |
| SLC8A1 | Calcium signaling pathway | ||
| THBS1 | Proteoglycans in cancer | ||
| TNF | MAPK signaling pathway | ||
| TNFRSF1B | Adipocytokine signaling pathway | ||
| ACSL1 | Adipocytokine signaling pathway | ||
| EDNRB | Calcium signaling pathway | ||
| miR-19b | Up | CALM1 | Calcium signaling pathway |
| TNF | Proteoglycans in cancer |
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal