Abstract
We used next generation sequencing (NGS) of the immunoglobulin genes to evaluate minimal residual disease (MRD) in 153 specimens from 32 patients with newly diagnosed adult B cell ALL enrolled in the phase II SWOG S0333 multi-center study.
We used the clonoSEQTM assay developed by Adaptive Biotechnologies that detects 1 leukemic cell in a background of 1 million nucleated cells and focuses in the B cell receptor (Ig). Initially, a set of pre-study specimens was sequenced in order to identify the precise sequence of the VDJ or DJ fragments. Clones representing more than 5% of the total repertoire of IgH molecules profiled were considered potentially leukemic. The follow-up specimen IgH repertoire sequences were compared to the diagnostic clonal ones and the leukemic marker sequence(s) previously identified were searched for explicitly.
At least one Ig clonotype was detected in 29/32 (91%) cases analyzed. The 3 remaining cases were reviewed, and in 2 cases the specimens available for NGS had been reported as having no blasts by morphology.
The leukemic clonal sequence was a complete VDJ rearrangement in 17/32 patients (53%), an incomplete DJ rearrangement in 8/32 patients (25%), and in 3/32 cases both VDJ and DJ rearrangements coexisted. One patient had a kappa light chain rearrangement. 17/32 (53%) cases contained more than one IgH rearrangement at diagnosis (median=2, range: 1 - 4). One of our patients is a potential case of therapy driven clonal selection. He presented at diagnosis with two related clones, one representing 75% of VDJ sequences and the second one 18%. At relapse, the second clone was responsible for most of the VDJ sequences (95%).
The NGS results were compared to the MRD results detected by multiparameter flow cytometry (MFC) in 66 specimens analyzed by both methods. The concordance between the methods in the qualitative determination of the presence or absence of leukemia was 82% (54/66). In 12 specimens (18%) MRD was detected by sequencing but not by MFC. One specimen had MRD detected at very low levels by MFC and was negative by NGS.
Our study includes 54 paired bone marrow (BM) and peripheral blood (PB) specimens. The median values of leukemia detected by NGS were 6-fold higher in BM than PB (range: 0.38 - 821-fold). Twenty-five pairs show no detectable MRD in either specimen. MRD was still detectable in 20 of the remaining 29 PB cases (for one of the pairs the BM specimen was negative). In 6/9 (67%) pairs of samples with disease detectable in BM but not in PB by NGS, no MRD was detected by MFC in the BM specimen.
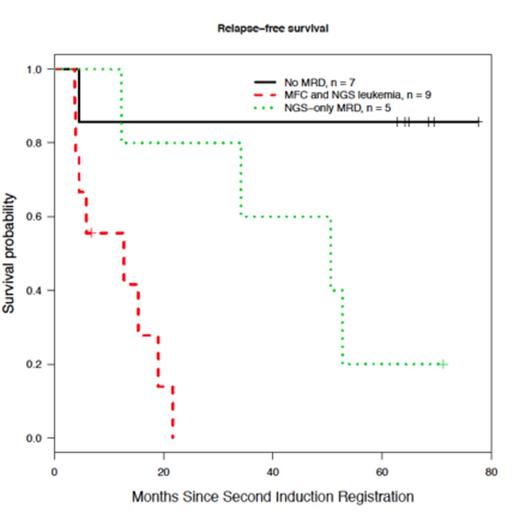
Lastly, outcome analysis was conducted in 21/32 patients with specimen available for MRD studies at the time of registration to second induction. Patients without MRD by NGS had a 5-year relapse free survival (RFS) of approximately 80%, while patients with MRD positive by both NGS and flow have the poorest outcome (p = 0.003) (see Figure). Patients with MRD detectable only by NGS have and intermediate RFS (p = 0.078, and p = 0.04 when compared to patients with MRD negative by both techniques, and patients with leukemia detected both by NGS and flow respectively). These results suggest that MRD detection by immunoglobulin gene sequencing is a very sensitive technique, and may identify patients with an excellent survival. Moreover, the increased sensitivity of the method may allow peripheral blood testing to supplement routine marrow sampling for MRD determination.
Williamson:Adaptive Biotechnologies: Employment, Equity Ownership. Kirsch:Adaptive Biotechnologies: Employment, Equity Ownership. Robins:Adaptive Biotechnologies: Consultancy, Equity Ownership.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal