Abstract
B-cell precursor acute lymphoblastic leukemia (BCP-ALL) is one of the most common cancers in children. The relapse incidence varies between 15% and 25%, dependent on the treatment protocol. It has been demonstrated that the presence of mutations in the RAS pathway genes, which regulate signal transduction upon binding of ligand to a variety of membrane receptors, are frequently associated with high-risk ALL and may be relapse drivers (Zhang et al., Blood 2011 118:3080-7; Irving et al., ASH 2013). Relapses in ALL are thought to result from the outgrowth of therapy-resistant residual leukemia cells and recent studies have shown a complex, dynamic architecture of clonal diversity in ALL (Anderson et al., Nature 2011 469:356-61; Notta et al. Nature 2011 496:362-7). In this study, we have investigated the clonal origin of RAS pathway mutations in relapsed BCP-ALL.
We screened 146 relapse samples from children with relapsed BCP-ALL for mutations in five RAS pathway genes using IonTorrent sequencing with a read-depth of approximately 150x. A total of 30% of relapse samples carried RAS pathway mutations, including mutations in KRAS (n=22), NRAS (n=13), PTPN11 (n=8), and FLT3 (n=2). No RAS mutations were found in 103 patients, whereas one patient had both a PTPN11 and a KRAS mutation, and another patient had two KRAS mutations. For 28 mutations we collected matched diagnosis samples and determined mutation presence using Sanger sequencing. We found that 12 of the RAS mutations were also present at diagnosis (43%) and 16 were initially not detected at diagnosis (57%). In order to gain more insight into the clonal evolution of relapse development, we performed amplicon based ultra-deep sequencing on the diagnosis samples, with an average read-depth of 50,000x for each mutation. The ultra-deep sequencing allows for sensitive and accurate detection of relapse-prone clones at diagnosis. A total of 22 mutations were identified at diagnosis samples (79%), of which 10 mutations had a low mutant allele frequency (average 3.8%), and were initially missed by Sanger sequencing (Fig.1). The 12 mutations determined using Sanger sequencing were detected at an average mutant allele frequency of 30.8%. We were unable to detect 6 mutations in the matched diagnosis samples, indicating that these mutations were newly acquired in the relapse clone at a time point after relapse. Our results indicate that cells harboring RAS pathway mutations are recurrently present in subclones at diagnosis. These cells may survive initial therapy and subsequently emerge at relapse. Patients with (minor clones with) RAS pathway mutations identified by ultra-deep sequencing, may benefit from treatment with MEK inhibitors added to the frontline therapy strategy.
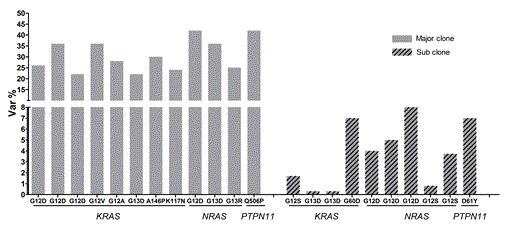
Backtracking of RAS pathway mutations by ultra-deep sequencing. A total of 22 mutations (out of 28) were detected in matched diagnosis samples. The 12 mutations (solid grey) determined by Sanger sequencing were detected at high mutant allele frequency ranging from 22% to 42%. Ten mutations (diagonally striped) were missed by Sanger sequencing but detected by ultra-deep sequencing. These mutations showed a low mutant allele frequency varying from 0.3% to 8%.
Backtracking of RAS pathway mutations by ultra-deep sequencing. A total of 22 mutations (out of 28) were detected in matched diagnosis samples. The 12 mutations (solid grey) determined by Sanger sequencing were detected at high mutant allele frequency ranging from 22% to 42%. Ten mutations (diagonally striped) were missed by Sanger sequencing but detected by ultra-deep sequencing. These mutations showed a low mutant allele frequency varying from 0.3% to 8%.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal