Abstract
Background: Although molecular complete remission (mCR) in multiple myeloma (MM) can be assessed by allele-specific oligonucleotide (ASO)-PCR, this technique requires preparation of clonotype-specific primers for each individual, which is laborious and time-consuming. We utilized the LymphoSIGHTTM platform, which employs consensus primers and next-generation sequencing (NGS) to amplify and sequence all rearranged immunoglobulin gene segments present in a myeloma clone, to assess mCR. This technique has been shown to have 1-2 logs greater sensitivity compared to ASO-PCR and flow cytometry, respectively (Faham et al, Blood 2012). Usage of the sequencing method for minimal residual disease (MRD) detection in MM may provide increased sensitivity and specificity, while overcoming the challenges associated with ASO-PCR. We compared the LymphoSIGHTTM method with ASO-qPCR for MRD detection in autografts and bone marrow (BM) in the autologous peripheral blood stem cell (PBSC) transplantation (ASCT) setting.
Methods: One hundred and nine Japanese patients with newly diagnosed MM who received various induction regimens prior to ASCT were retrospectively analyzed. All patients had achieved a partial response (PR) or better after ASCT. BM slides from 84 MM patients and fresh/frozen BM cells from 25 MM patients at diagnosis, as well as autografts/post-ASCT BM cells from each patient, were obtained for DNA extraction. IGH-based ASO-qPCR was performed as described previously (Methods Mol Biol 2009). Using universal primer sets, we amplified IGH variable (V), diversity (D), and joining (J) gene segments, IGH-DJ, and IGK from genomic DNA. Amplified products were subjected to deep sequencing using NGS. Reads were analyzed using standardized algorithms for clonotype determination. Myeloma-specific clonotypes were identified for each patient based on their high frequency in BM samples.
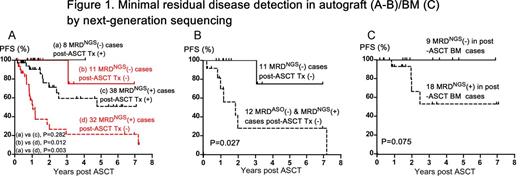
Results : Myeloma clonotypes could be identified in autografts/post-ASCT BM cells in 98 of 109 patients (90%) and by ASO-qPCR in 63 of 101 patients (62%). MRD by NGS was assessed in autografts of 89 patients. 70 of 89 patients (79%) were positive by NGS; 28 of 62 patients (45%) were positive by ASO-qPCR. Although we observed a high correlation between NGS and PCR MRD results at MRD levels of 10-5 or higher, the sensitivity of ASO-PCR was 10-4-10-5, whereas that of NGS was 10-6 or lower when a sufficient amount of DNA was available for analysis. Eight cases where MRD was not detected in the autograft by NGS (MRDNGS(-)) and 38 MRDNGS(+) cases received post-ASCT therapy using novel agents such as bortezomib/lenalidomide/thalidomide, while 11 MRDNGS(-) cases and 32 MRDNGS(+) cases were followed without post-ASCT therapy. The MRDNGS(-) cases without post-ASCT therapy showed significantly better progression-free survival (PFS) than the MRDNGS(+) cases without post-ASCT therapy (P = 0.012) (Figure 1A) although overall survival rates were comparable between these groups. To investigate the value of sensitive detection by NGS, we compared PFS in 11 MRDNGS(-) cases (Group 1) with the 12 MRDNGS(+) cases where MRD was not detected by ASO-qPCR (MRDASO(-)) (Group 2). The patients in both groups did not receive any post-ASCT therapy. Group 1 showed significantly better PFS than Group 2 (P = 0.027) (Figure 1B). Furthermore, 9 MRDNGS(-) in post-ASCT BM cases tended to show a better PFS than 18 MRDNGS(+) in post-ASCT BM cases (P = 0.075) (Figure 1C). In a multivariate analysis, post-ASCT therapy using novel agents (P <0.001) and MRDNGS(-) in autograft (P=0.025) were independently associated with superior PFS while ISS I/II vs III (P = 0.387) and MRDASO(-) in autograft (P=0.174) were not.
Conclusions: In this study, we showed the prognostic value of MRD detection using the NGS-based LymphoSIGHT platform in autografts of patients with MM who received and in those who did not receive post-ASCT therapy with novel agents. The NGS platform has improved sensitivity compared with ASO-qPCR in detecting MRD in autografts. Patients with low level MRD detected by NGS but not by ASO-qPCR have worse prognosis compared to patients who are MRD negative by sequencing, which underscores the need for sensitive detection.
Zheng:Sequenta, Inc.: Employment. Moorhead:Sequenta, Inc.: Employment. Faham:Sequenta, Inc.: Employment, Equity Ownership, Membership on an entity's Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal