Key Points
DPY30 is important for the proliferation and proper differentiation of human hematopoietic progenitor cells.
dpy30 and efficient H3K4 methylation are essential for the normal hematopoiesis of zebrafish.
Abstract
Epigenetic mechanisms, including histone modifications, have emerged as important factors influencing cell fate determination. The functional role of H3K4 methylation, however, remains largely unclear in the maintenance and differentiation of hematopoietic stem cells (HSCs)/hematopoietic progenitor cells (HPCs). Here we show that DPY30, a shared core subunit of the SET1/MLL family methyltransferase complexes and a facilitator of their H3K4 methylation activity, is important for ex vivo proliferation and differentiation of human CD34+ HPCs. DPY30 promotes HPC proliferation by directly regulating the expression of genes critical for cell proliferation. Interestingly, while DPY30 knockdown in HPCs impaired their differentiation into the myelomonocytic lineage, it potently promoted hemoglobin production and affected the kinetics of their differentiation into the erythroid lineage. In an in vivo model, we show that morpholino-mediated dpy30 knockdown resulted in severe defects in the development of the zebrafish hematopoietic system, which could be partially rescued by coinjection of dpy30 messenger RNA. Taken together, our results establish a critical role of DPY30 in the proliferation and appropriate differentiation of hematopoietic progenitor cells and in animal hematopoiesis. Finally, we also demonstrate a crucial role of DPY30 in the growth of several MLL1-fusion–mediated leukemia cell lines.
Introduction
The maintenance, proliferation, and differentiation of stem and progenitor cells are ultimately controlled at the level of gene expression, which is closely tied to the global and local epigenetic status in the cell. A paradigm for such epigenetic control of gene expression is shown by 2 well-established antagonistic histone modifications: H3K27 methylation, catalyzed by the Polycomb group complexes, and H3K4 methylation, mainly catalyzed by the Trithorax group complexes.1 Although H3K27 methylation is generally associated with gene repression, H3K4 methylation is prevalently associated with gene activation.2,3 Roles for Polycomb group complexes and H3K27 methylation have been extensively studied in both embryonic stem cells (ESCs)4-6 and hematopoietic stem cells (HSCs) and hematopoietic progenitor cells (HPCs).7-15 On the other hand, the functional roles for H3K4 methylation in the maintenance and differentiation of stem and progenitor cells remain largely unclear.
The SET1/MLL family complexes are the most notable H3K4 methyltransferases in mammals. They are composed of either SET1A, SET1B, MLL1, MLL2, MLL3, or MLL4 as the catalytic subunit, and WDR5, RBBP5, ASH2L, and DPY30 as integral core subunits that are required for the full methylation activity of these complexes.2,16-19 The functional roles of the SET1/MLL complexes are especially pertinent to the hematopoietic system, as MLL1 (mixed lineage leukemia [MLL]) is a common target of chromosomal translocations in human acute leukemias,20-22 and other subunits of the complexes are extensively associated with diseases such as blood cancers.23,24 The molecular mechanisms underlying MLL1-fusion–mediated leukemogenesis remain incompletely understood.25-27 Mll1 is essential for normal hematopoiesis28-32 and a wild-type Mll1 allele is critical for MLL1-AF9–mediated leukemogenesis.33 Although the H3K4 methylation activity of MLL1 was concluded in a recent report to be important for MLL1-AF9–mediated leukemogenesis,34 it was later shown in another report to be dispensable for this process and normal hematopoiesis,35 underscoring the complex relationship of the enzymatic activity and the function of the protein. The lack of impact on global or gene-specific H3K4 methylation upon acute Mll1 deletion35 makes it difficult to investigate the role of H3K4 methylation in hematopoiesis through Mll1 deletion. Hematopoietic studies on other SET1/MLL complex subunits are scarce and have limited information on the involvement of the methylation activity,36 thus leaving a major gap between chromatin regulation by H3K4 methylation and hematopoiesis.
We have previously shown that the DPY30 subunit of the SET1/MLL complexes is important for facilitating genome-wide H3K4 methylation.37 Although dispensable for self-renewal of mouse ESCs, Dpy30 is crucial for induction of developmental genes and efficient differentiation of ESCs.37 It remains unknown whether DPY30 plays a similar role in the maintenance and differentiation of somatic stem and progenitor cells. Here we sought to address a fundamental question regarding the role of H3K4 methylation in hematopoietic progenitor function by depleting DPY30 in various systems including human HPCs and zebrafish.
Methods
Purification, culture, and infection of human CD34+ cells and leukemia cells
Mononuclear cells were isolated from mobilized peripheral blood from healthy donors with their informed consent. CD34+ cells were purified by positive selection using a MACS separator, LS+ column, and CD34 Micro Bead Kit (Miltenyi Biotec, Auburn, CA), and were verified to be >97% pure by flow cytometry analysis. An additional batch of purified CD34+ cells was purchased from the Cincinnati Children’s Hospital Medical Center. CD34+ cells were cultured in Iscove modified Dulbecco medium ([IMDM]; Invitrogen, Grand Island, NY) containing 20% BIT 9500 Serum Substitute (cat.# 09500; STEMCELL Technologies, Vancouver, BC, Canada) freshly supplemented with stem cell factor (SCF) (100 ng/mL), Fms-like tyrosine kinase 3 (FLT-3) (10 ng/mL), thrombopoietin (TPO) (100 ng/mL), IL-6 (20 ng/mL), and low density lipoprotein (LDL) (40 ng/mL). The cytokines were purchased from PeproTech (Rocky Hill, NJ). K562, HEL, and MOLM-13 cells (gift of Xinyang Zhao), THP-1 cells (gift of Jianjun Chen), RS4;11 cells (gift of William Placzek), KOPN-8 cells (gift of Charles Hemenway), TF-1 cells (ATCC, Manassas, VA), and MEL-745A cells38 were cultured in RPMI-1640 (Invitrogen) containing 10% fetal bovine serum (Invitrogen). TF-1 culture also contained 2 ng/mL granulocyte macrophage–colony-stimulating factor (GM–CSF).
pLKO.1-based lentiviral constructs expressing scramble control (Addgene plasmid 1864) and DPY30 short hairpin (shRNAs) (supplemental Table 1, available on the Blood Web site) were from Addgene and OpenBiosystems, respectively. Viral particles were produced according to the recommended protocols (Addgene) and concentrated when necessary. After 1 to 3 days of growth, CD34+ cells were infected with concentrated viruses in the presence of 8 µg/mL of polybrene (Sigma-Aldrich, St. Louis, MO). Two days after the infection, puromycin (2 µg/mL) was added, and after 3 more days, the cells were used for various assays including the liquid differentiation assays. Leukemia cells were infected with unconcentrated viruses with the same method, except that 8 µg/mL of protamine sulfate (Sigma-Aldrich) was used for infecting KOPN-8 cells. 5-bromo-2′-deoxyuridine 5′-triosphate (BrdUTP) incorporation and cell apoptosis were analyzed on the stably infected cells using the fluorescein isothiocyanate (FITC) BrdU flow kit and FITC Annexin V apoptosis detection kit (both from BD Biosciences, Franklin Lakes, NJ) with manufacturer recommended protocols.
Gene expression and ChIP assays
Details of the gene expression and chromatin immunoprecipitation (ChIP) assays are provided in the supplemental Methods. The microarray data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE59643).
Methylcellulose colony assays
Indicated numbers of stably infected live CD34+ cells were plated in human methylcellulose-enriched medium (HSC005; R&D Systems, Minneapolis, MN) containing erythropoietin (EPO) (3 IU/mL), granulocyte CSF (G-CSF) (20 ng/mL), GM-CSF (20 ng/mL), IL-3 (20 ng/mL), IL-6 (20 ng/mL). and SCF (50 ng/mL). Burst-forming units-erythroid (BFU-E), granulocyte colony-forming units (CFU-G), granulocyte-macrophage CFU (CFU-GM), and granulocyte erythrocyte macrophage megakaryocyte CFU (CFU-GEMM) were scored 14 days later. A total of 10 000 stably infected live MLL1-fusion–mediated leukemia cells were plated in human methylcellulose base media (HSC002; R&D Systems) with no cytokines. After recovery of colonies from the first plating, 10 000 live cells were replated in the same medium. No antibiotics were contained in the methylcellulose media.
Differentiation assays in liquid culture
Puromycin-selected CD34+ cells were cultured in medium that supports either erythroid (IMDM, 20% BIT 9500, SCF 100 ng/mL, EPO 1 units/mL) or myelomonocytic differentiation (IMDM, 20% BIT 9500, SCF 100 ng/mL, FLT-3 ligand 10 ng/mL, IL-3 20 ng/mL, IL-6 20 ng/mL, GM-CSF 20 ng/mL, G-CSF 20 ng/mL). All differentiation media contained 2 µg/mL puromycin. At indicated days after culture under differentiation conditions, cells were stained with Glycophorin A-PE (BD 561051) and CD71-APC (BD 561940) for erythroid differentiation and with CD34-APC (BD 560940) and CD11b-PE (BD 561001) for myeloid differentiation. Flow cytometry was performed using LSRFortessa Cell Analyzer (BD Biosciences) and the data were analyzed by FlowJo (Tree Star, Ashland, OR).
Zebrafish work
Zebrafish (Danio rerio, strain AB) were maintained under standard conditions in the University of Alabama at Birmingham (UAB) zebrafish core facility in accordance with the UAB policy on animal care and use. Antisense morpholino oligonucleotides (MOs) were purchased from Gene Tools, LLC (Philomath, OR). Indicated dose of MO at 0.5 to 1 nanoliter was microinjected into embryos at the 1 to 2 cell stage. Zebrafish dpy30 messenger RNA (mRNA) was injected alone or coinjected with MO at 100 picogram/nanoliter (50-100 picogram/embryo). Details of zebrafish work are provided in the supplemental Methods.
Results
DPY30 directly promotes ex vivo proliferation of primary hematopoietic progenitors
To investigate the role of DPY30 in human HPCs, we used 2 different shRNA-expressing lentiviruses to stably and efficiently knock down (KD) DPY30 expression (Figure 1A) in primary human CD34+ cells purified from mobilized peripheral blood, which are enriched for HPCs. We confirmed that global H3K4 methylation was reduced by DPY30 KD (Figure 1B). As shown by colony-forming–cell assays, DPY30 KD by either shRNA significantly reduced the colony numbers for various types of myeloid lineage cells (Figure 1C and supplemental Figure 2A). The reduction was most striking for CFU-GEMM, which are derived from immature multilineage progenitors (Figure 1C and supplemental Figure 2A), and BFU-E (Figure 1C and supplemental Figures 1 and 2A), which are derived from immature erythroid progenitors. Interestingly, the BFU-E colonies that did form from the DPY30-KD background typically appeared to be uniformly larger and redder on average when compared with the control BFU-E colonies (supplemental Figure 1). Consistent with the reduced number of functional progenitors after DPY30 KD, CD34+ cell proliferation, as indicated by the BrdUTP incorporation into newly synthesized DNA, was also found to be significantly reduced by DPY30 KD (Figure 1D and supplemental Figure 2B).
DPY30 KD impaired the proliferation of human HPCs. Human CD34+ cells were depleted of DPY30 by 2 different shRNAs (#2 and #5). (A) The KD efficiency was determined by qRT-PCR and normalized against ACTB. Averages ± standard deviation (SD) from 5 independent biological repeats (infections) are plotted. (B) Effect of DPY30 KD (short hairpin [sh]#2 and #5) on global H3K4 methylation level in cells from 2 different sources was determined by western blot analysis. H3K4me1, H3K4me2, and H3K4me3: H3K4 mono-, di-, and tri-methylation, respectively. (C) Various types of myeloid colonies were identified and quantified after the colony-forming–cell assay using control or DPY30-KD cells. Averages ± SD from 3 assays for 1 of 4 independent biological repeats are plotted (the rest are shown in supplemental Figure 2A). P < .01 (Student t test) between the control and either DPY30 shRNAs for all types of colonies. (D) Proliferation of CD34+-gated cells was monitored by BrdU incorporation after culture in the presence of BrdUTP for 12 hours, and the percentage of BrdU-positive cells was quantified from 3 independent biological repeats. *P < .05 (Student t test). (E) Global gene expression was examined by microarray, followed by gene ontology analysis on the genes downregulated by both shRNAs to below three-fourths of the levels in control cells (listed in supplemental Table 3). (F) Expression of selected genes from microarray assays was examined by qRT-PCR against POLR2A. Two different sets of primers were used for MYC. Averages ± SD from 4 independent biological repeats are plotted. P < .05 (Student t test) between control and either DPY30 shRNAs for all genes. (G) Chromatin immunoprecipitation assays for total H3, H3K4me3, and DPY30 binding at the transcription start sites of a negative control gene (OR2J3) and those genes examined in panel F in human CD34+ cells. Averages ± SD from triplicate assays are plotted.
DPY30 KD impaired the proliferation of human HPCs. Human CD34+ cells were depleted of DPY30 by 2 different shRNAs (#2 and #5). (A) The KD efficiency was determined by qRT-PCR and normalized against ACTB. Averages ± standard deviation (SD) from 5 independent biological repeats (infections) are plotted. (B) Effect of DPY30 KD (short hairpin [sh]#2 and #5) on global H3K4 methylation level in cells from 2 different sources was determined by western blot analysis. H3K4me1, H3K4me2, and H3K4me3: H3K4 mono-, di-, and tri-methylation, respectively. (C) Various types of myeloid colonies were identified and quantified after the colony-forming–cell assay using control or DPY30-KD cells. Averages ± SD from 3 assays for 1 of 4 independent biological repeats are plotted (the rest are shown in supplemental Figure 2A). P < .01 (Student t test) between the control and either DPY30 shRNAs for all types of colonies. (D) Proliferation of CD34+-gated cells was monitored by BrdU incorporation after culture in the presence of BrdUTP for 12 hours, and the percentage of BrdU-positive cells was quantified from 3 independent biological repeats. *P < .05 (Student t test). (E) Global gene expression was examined by microarray, followed by gene ontology analysis on the genes downregulated by both shRNAs to below three-fourths of the levels in control cells (listed in supplemental Table 3). (F) Expression of selected genes from microarray assays was examined by qRT-PCR against POLR2A. Two different sets of primers were used for MYC. Averages ± SD from 4 independent biological repeats are plotted. P < .05 (Student t test) between control and either DPY30 shRNAs for all genes. (G) Chromatin immunoprecipitation assays for total H3, H3K4me3, and DPY30 binding at the transcription start sites of a negative control gene (OR2J3) and those genes examined in panel F in human CD34+ cells. Averages ± SD from triplicate assays are plotted.
Then we analyzed the effect of DPY30 KD on global gene expression in these cells by microarray analyses. As shown in supplemental Table 3, 284 genes were downregulated by both shRNAs to below three-quarters, and 62 genes to below two-thirds, of the levels in control cells. Gene ontology analysis revealed that genes involved in DNA replication and cell cycle regulation were mostly overrepresented among these genes (Figure 1E and supplemental Figure 2C). We confirmed the microarray results for selected genes including MYC, CDCA7, MCM2, MCM3, and MCM4, using quantitative polymerase chain reaction (qPCR) (Figure 1F). As shown by chromatin immunoprecipitation assays, the transcription start sites of these tested genes, but not that of a negative control gene OR2J3, were highly enriched for H3K4me3 and DPY30 (Figure 1G). These results suggest that DPY30 directly promotes expression of genes critically involved in DNA replication and cell cycle progression in human HPCs.
DPY30 KD impairs myelomonocytic differentiation of human HPCs
Having established an important role of DPY30 in the ex vivo proliferation of HPCs, we asked whether DPY30 has a role in HPC differentiation. Control and DPY30-KD HPCs were assayed in liquid culture under the myelomonocytic differentiation condition. At the later stage of myelomonocytic differentiation, a significant portion of the control cells, but a much smaller portion of the DPY30-KD cells, exhibited a macrophage-like morphology, with irregular cell shapes and some adherent to the bottom of the well (Figure 2A and supplemental Figures 3 and 4). Such morphologic difference suggests that DPY30 KD impairs the myelomonocytic differentiation into macrophages. Consistent with the morphologic effect, DPY30 KD modestly, but consistently, impaired the differentiation into CD34−CD11b+ cells at the mid and later time points during the assays (Figure 2B and supplemental Figure 5A). Moreover, more residual undifferentiated CD34+CD11b− cells were seen for DPY30-KD cells after several days of differentiation (supplemental Figure 5A-B). Cell proliferation was not found to be significantly reduced by DPY30 KD during the myelomonocytic differentiation (supplemental Figure 6).
DPY30 KD impaired myelomonocytic differentiation of human HPCs. Control and DPY30-KD human CD34+ cells were cultured with appropriate cytokine conditions that permit myelomonocytic differentiation. (A) Cell morphology at day 11 under such culture condition. (B) Percentage of myelomonocytic population (CD34−CD11b+) at day 14 in the differentiation assays were quantified. Averages ± SD from 3 biological repeats (infections) are plotted. **P < .01 (Student t test). (C) RNAs from the control and DPY30-KD (by short hairpin [sh]#2) cells on day 9 during myelomonocytic differentiation were subject to microarray assays followed by gene ontology analysis for the genes downregulated over twofold by DPY30 KD (listed in supplemental Table 4). (D) RNAs from the control and DPY30-KD (by sh#2 and #5) cells at day 0 (d0) and day 14 (d14) during myelomonocytic differentiation were subject to quantitative reverse-transcription polymerase chain reaction for selected genes highlighted in supplemental Table 4. Averages ± SD from 2 independent biological repeats are plotted.
DPY30 KD impaired myelomonocytic differentiation of human HPCs. Control and DPY30-KD human CD34+ cells were cultured with appropriate cytokine conditions that permit myelomonocytic differentiation. (A) Cell morphology at day 11 under such culture condition. (B) Percentage of myelomonocytic population (CD34−CD11b+) at day 14 in the differentiation assays were quantified. Averages ± SD from 3 biological repeats (infections) are plotted. **P < .01 (Student t test). (C) RNAs from the control and DPY30-KD (by short hairpin [sh]#2) cells on day 9 during myelomonocytic differentiation were subject to microarray assays followed by gene ontology analysis for the genes downregulated over twofold by DPY30 KD (listed in supplemental Table 4). (D) RNAs from the control and DPY30-KD (by sh#2 and #5) cells at day 0 (d0) and day 14 (d14) during myelomonocytic differentiation were subject to quantitative reverse-transcription polymerase chain reaction for selected genes highlighted in supplemental Table 4. Averages ± SD from 2 independent biological repeats are plotted.
As cell identity is ultimately determined by its gene expression profile, we performed microarray analyses to characterize the effect of DPY30 KD on gene expression after myelomonocytic differentiation. Our results indicate that the genes whose postdifferentiation expression was reduced over twofold by DPY30 KD (235 genes listed in supplemental Table 4) were significantly enriched for defense response, antigen processing and presentation, and inflammatory response (Figure 2C), hallmark functions for differentiated myelomonocytic cells. Such enrichment was specific, as it was not found for genes whose expression was enhanced by DPY30 KD (data not shown). By quantitative reverse-transcription PCR (qRT-PCR), we further showed that while these genes were efficiently induced in control cells after culture under myelomonocytic differentiation condition, their induction was greatly impaired in the DPY30-KD cells (Figure 2D and supplemental Figure 5C). Taken together, these results demonstrate an important role for DPY30 in the myelomonocytic differentiation of human HPCs.
DPY30 KD promotes erythroid maturation in HPCs and erythroleukemia cells
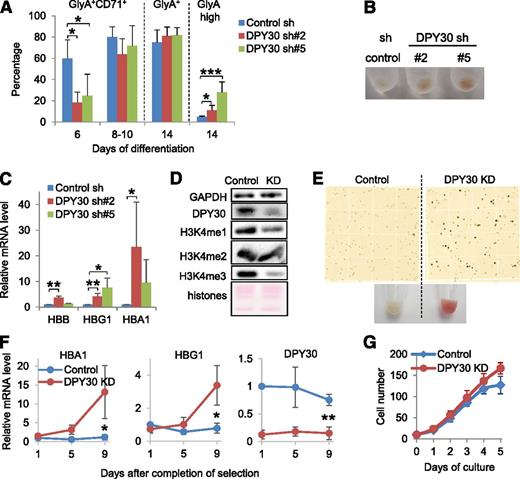
Next we examined the effect of DPY30 KD on erythroid differentiation of human HPCs using a differentiation assay in liquid culture containing EPO. We first showed that DPY30 KD had little effect on cell proliferation (supplemental Figure 6) and overall cell morphology (supplemental Figure 7) during the erythroid differentiation assays. Flow cytometry analyses of Glycophorin A (GlyA) and CD71 expression showed an initial decrease in the generation of the mature erythroid precursors (GlyA+CD71+) for the DPY30-KD cells (day 6 in Figure 3A and supplemental Figure 8). However, erythroid differentiation was consistently enhanced in DPY30-KD cells to a similar level as in control cells at the mid- and late stages of the assays (days 8-10 and 14 in Figure 3A and supplemental Figure 8). By the end of the differentiation process, DPY30-KD cells contained a significantly higher percentage of the GlyA-high population (day 14 in Figure 3A and supplemental Figure 8) and displayed a markedly enhanced red color after pelleting compared with the control cells (Figure 3B). Furthermore, hemoglobin genes were strongly upregulated upon DPY30 KD before exposure to EPO-containing differentiation medium (Figure 3C), suggesting an intrinsic repressive role of DPY30 in the expression of hemoglobin genes in human HPCs.
DPY30 KD promoted erythroid maturation of human HPCs and erythroleukemia cells. Human CD34+ cells were depleted of DPY30 by 2 different shRNAs (#2 and #5), and were induced to differentiate into erythroid lineage in liquid culture with appropriate cytokine conditions (containing 1 ug/mL EPO). (A) Indicated cell surface markers were analyzed and quantified by flow cytometry at different days during differentiation. Averages ± SD from 4 independent biological repeats are plotted. *P < .05; ***P < .001 (Student t test). (B) Image of pelleted cells on day 14 of erythroid differentiation. (C) The expression of hemoglobin genes before the differentiation assays was determined by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Averages ± SD from 4 biological repeats are plotted. *P < .05; **P < .01 (Student t test). (D-E) Ten days after DPY30 KD (by sh#2 viral infection), K562 cells were checked by western blot analysis for KD and effect on H3K4 methylation (D), and stained by benzidine (E, top) and pelleted for image (E, bottom). (F) Expression of HBA1, HBG1, and DPY30 in control and DPY30-KD K562 cells was monitored by qRT-PCR against ACTB. Completion of selection, indicated by loss of viability of most uninfected cells after culturing in the presence of puromycin, was 4 days after viral infection. Averages ± SD from 3 independent biological repeats are plotted. *P < .05; **P < .01 (Student t test). (G) Growth of control and DPY30-KD K562 cells was monitored daily. Averages ± SD from 3 biological repeats are plotted.
DPY30 KD promoted erythroid maturation of human HPCs and erythroleukemia cells. Human CD34+ cells were depleted of DPY30 by 2 different shRNAs (#2 and #5), and were induced to differentiate into erythroid lineage in liquid culture with appropriate cytokine conditions (containing 1 ug/mL EPO). (A) Indicated cell surface markers were analyzed and quantified by flow cytometry at different days during differentiation. Averages ± SD from 4 independent biological repeats are plotted. *P < .05; ***P < .001 (Student t test). (B) Image of pelleted cells on day 14 of erythroid differentiation. (C) The expression of hemoglobin genes before the differentiation assays was determined by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). Averages ± SD from 4 biological repeats are plotted. *P < .05; **P < .01 (Student t test). (D-E) Ten days after DPY30 KD (by sh#2 viral infection), K562 cells were checked by western blot analysis for KD and effect on H3K4 methylation (D), and stained by benzidine (E, top) and pelleted for image (E, bottom). (F) Expression of HBA1, HBG1, and DPY30 in control and DPY30-KD K562 cells was monitored by qRT-PCR against ACTB. Completion of selection, indicated by loss of viability of most uninfected cells after culturing in the presence of puromycin, was 4 days after viral infection. Averages ± SD from 3 independent biological repeats are plotted. *P < .05; **P < .01 (Student t test). (G) Growth of control and DPY30-KD K562 cells was monitored daily. Averages ± SD from 3 biological repeats are plotted.
The seemingly complicated effect of DPY30 KD on erythroid progenitors prompted further studies in cells with a relatively more restricted fate, for instance, human and mouse erythroleukemia cells. DPY30 KD in K562 cells, a human erythroleukemia cell line, reduced the global H3K4 methylation level (Figure 3D), potently drove hemoglobin production, as evident by the striking red color of the pelleted cells (Figure 3E), the dramatically increased benzidine-positive cells (Figure 3E), and the upregulated hemoglobin gene expression (Figure 3F). Furthermore, although GlyA was already expressed at a high level in control cells, it was further upregulated after DPY30 KD (supplemental Figure 9A). Importantly, proliferation was not reduced by DPY30 KD (Figure 3G), indicating that the increased erythroid differentiation was not a result of reduced cell proliferation. Consistent with the effects in K562 cells, DPY30 KD in 2 other human erythroleukemia cell lines, TF-139 and HEL,40 significantly enhanced hemin-induced hemoglobin production (supplemental Figure 9B). Furthermore, Dpy30 KD in MEL-745A,41,42 a mouse erythroleukemia cell line, promoted their spontaneous as well as DMSO-induced differentiation toward erythrocytes in a dose-dependent manner, as shown by an increase in benzidine-positive cells (supplemental Figure 9C) and an upregulation of mouse hemoglobin genes (supplemental Figure 9D). These results corroborate those from the EPO-directed erythroid differentiation of human HPCs in suggesting that DPY30 KD promotes erythroid maturation.
DPY30 ortholog is required for embryonic hematopoiesis in zebrafish
Next we examined a role of DPY30 in a live vertebrate model, zebrafish, which has a highly conserved DPY30 ortholog, dpy30 (Figure 4A). Two different translation-blocking MOs were designed to target the zebrafish dpy30 transcript at the 5′ (MO-1) or 3′ (MO-2) to the translation start codon (Figure 4B). Injection of 3.5 to 7.0 ng (designated as 1× dose) of a nontargeting control MO generated no detectable abnormalities compared with uninjected embryos (supplemental Figure 10A and supplemental Video 1), and we therefore used uninjected embryos as the control for most of the later experiments. Injection of either of the 2 dpy30 MOs at lower doses consistently resulted in severe developmental abnormalities. The most obvious phenotypes were cardiac edema (not shown) and reduction or abrogation of blood circulation, as demonstrated by the time-lapse video microscopy (Figure 4C-D; supplemental Figures 10B and 11A, and supplemental Video 2). O-dianisidine staining for hemoglobin showed a severely reduced number of red blood cells in most of the morphant embryos (Figure 4E-F and supplemental Figures 10C and 11B). The abnormalities persisted as late as 5 days after fertilization (data not shown), suggesting that they were not merely due to a delay in development.
DPY30 ortholog is essential for hematopoiesis in zebrafish. (A) Alignment of the human DPY30 and zebrafish dpy30 proteins. Identical amino acids are denoted in bold type and asterisks indicate stop codons. (B) Design of zebrafish dpy30 MOs in relationship to the zebrafish dpy30 gene. The red and gray bars represent the dpy30 MOs. (C and D) Effect of dpy30 MOs on zebrafish blood circulation. (C) Representative images of circulating blood cells (picture frames from the time-lapse video microscopy) in uninjected control zebrafish and zebrafish injected with 0.125× dose of dpy30 MO-1. Green arrows point at the blood cells. (D) Quantification of blood circulation status at indicated time (after fertilization) in uninjected control zebrafish (Control), zebrafish injected with 0.25× dose of dpy30 MO-1 (MO-1), with 0.25× dose of dpy30 MO-1 plus 50 to 100 pg of dpy30 mRNA (MO-1 + mRNA), 1× dose of control MO (cMO), or 0.375× dose of dpy30 MO-2 (MO-2). (E and F) Effect of dpy30 MOs on zebrafish hematopoiesis examined by o-dianisidine staining at 48 hpf. Zebrafish were uninjected (Control), injected with 0.25× dose of dpy30 MO-1 (MO-1), with 0.25× dose of dpy30 MO-1 plus 50 to 100pg of dpy30 mRNA (MO-1 + mRNA), or with 0.375× dose of dpy30 MO-2 (MO-2). (E) Typical results for o-dianisidine staining showing zebrafish with normal red blood cell production and with severe or moderate anemia. Red arrows point at the red blood cells. Note that although some residual red blood cells could sometimes be visualized at the heart region, none was seen in the periphery of the severely anemic fish. These images, from top to bottom, were selected from the Control, MO-1, and MO-1 + mRNA groups, respectively. (F) O-dianisidine staining results were quantified for each category of red blood cell levels (according to the images shown in [E]). Plotted are averages ± SD from 3 independent biological repeats for the left graph (also shown individually in supplemental Figure 10C), and from 2 biological repeats for the right graph (also shown individually in supplemental Figure 11B). **P < .01; ***P < .001 (Student t test). (G) Western blot analysis was used for H3K4 methylation using total extracts from 48 hpf uninjected control zebrafish and zebrafish injected with 0.25× dose of dpy30 MO-1. (H) In situ hybridization for rag1 at day 5 after fertilization in uninjected control zebrafish and zebrafish injected with 0.375× dose of dpy30 MO-2. Dorsal (top) and lateral (bottom) views are shown. The arrow points at the rag1 signal. (I) In situ hybridization for tal1, gata1a, and fli1a at 30 hpf in uninjected control zebrafish and zebrafish injected with 0.25× dose of dpy30 MO-1.
DPY30 ortholog is essential for hematopoiesis in zebrafish. (A) Alignment of the human DPY30 and zebrafish dpy30 proteins. Identical amino acids are denoted in bold type and asterisks indicate stop codons. (B) Design of zebrafish dpy30 MOs in relationship to the zebrafish dpy30 gene. The red and gray bars represent the dpy30 MOs. (C and D) Effect of dpy30 MOs on zebrafish blood circulation. (C) Representative images of circulating blood cells (picture frames from the time-lapse video microscopy) in uninjected control zebrafish and zebrafish injected with 0.125× dose of dpy30 MO-1. Green arrows point at the blood cells. (D) Quantification of blood circulation status at indicated time (after fertilization) in uninjected control zebrafish (Control), zebrafish injected with 0.25× dose of dpy30 MO-1 (MO-1), with 0.25× dose of dpy30 MO-1 plus 50 to 100 pg of dpy30 mRNA (MO-1 + mRNA), 1× dose of control MO (cMO), or 0.375× dose of dpy30 MO-2 (MO-2). (E and F) Effect of dpy30 MOs on zebrafish hematopoiesis examined by o-dianisidine staining at 48 hpf. Zebrafish were uninjected (Control), injected with 0.25× dose of dpy30 MO-1 (MO-1), with 0.25× dose of dpy30 MO-1 plus 50 to 100pg of dpy30 mRNA (MO-1 + mRNA), or with 0.375× dose of dpy30 MO-2 (MO-2). (E) Typical results for o-dianisidine staining showing zebrafish with normal red blood cell production and with severe or moderate anemia. Red arrows point at the red blood cells. Note that although some residual red blood cells could sometimes be visualized at the heart region, none was seen in the periphery of the severely anemic fish. These images, from top to bottom, were selected from the Control, MO-1, and MO-1 + mRNA groups, respectively. (F) O-dianisidine staining results were quantified for each category of red blood cell levels (according to the images shown in [E]). Plotted are averages ± SD from 3 independent biological repeats for the left graph (also shown individually in supplemental Figure 10C), and from 2 biological repeats for the right graph (also shown individually in supplemental Figure 11B). **P < .01; ***P < .001 (Student t test). (G) Western blot analysis was used for H3K4 methylation using total extracts from 48 hpf uninjected control zebrafish and zebrafish injected with 0.25× dose of dpy30 MO-1. (H) In situ hybridization for rag1 at day 5 after fertilization in uninjected control zebrafish and zebrafish injected with 0.375× dose of dpy30 MO-2. Dorsal (top) and lateral (bottom) views are shown. The arrow points at the rag1 signal. (I) In situ hybridization for tal1, gata1a, and fli1a at 30 hpf in uninjected control zebrafish and zebrafish injected with 0.25× dose of dpy30 MO-1.
The phenotypes we observed are very different from the commonly observed nonspecific morpholino toxicity.43 To further exclude the possibility of off-target effects, we synthesized zebrafish dpy30 mRNA that lacked the MO-1 target sequence. Although injection of the dpy30 mRNA alone resulted in a slightly elevated mortality rates, its coinjection together with dpy30 MO-1, partially, but significantly rescued the severe defects in blood circulation (Figure 4D; supplemental Figure 10B and supplemental Video 3) and red blood cell production (Figure 4F and supplemental Figure 10C). In addition to erythroid development, we also examined lymphoid development by in situ hybridization assays for rag1, a gene specific and required for lymphopoiesis. In contrast to the normal expression of rag1 in the bilateral thymic lobes, rag1 signal was greatly reduced or abolished in the dpy30 morphant fish 5 days after fertilization (Figure 4H and supplemental Figure 12), indicating a defect in lymphoid development. Therefore, our results collectively indicate that dpy30 is crucial for embryonic hematopoiesis in zebrafish. Consistent with a conserved dpy30 function in facilitating H3K4 methylation, we found a clear defect in H3K4 methylation in the dpy30 morphant fish extract (Figure 4G). To examine the effects on HSC/HPC, we performed in situ hybridization for several hematopoiesis markers including tal1, gata1a, and fli1a, some of which are functionally critical for early hematopoiesis and/or HSC formation.44 Surprisingly, despite the defective erythropoiesis and lymphopoiesis, expression of all of these markers at 30 hours post fertilization (hpf) was enhanced in the morphant fish (Figure 4I and supplemental Figure 13), suggesting an abnormal accumulation of HSCs and/or early HPCs in the embryos with lowered levels of dpy30 and global H3K4 methylation.
DPY30 KD impairs the growth of MLL1-fusion–mediated leukemia cell lines
Having established a clear role of DPY30 in normal hematopoiesis, we sought to determine if it also plays a role in MLL-fusion-mediated leukemogenesis, especially considering a requirement of the wild-type MLL1 in MLL1-AF9–mediated leukemogenesis. We depleted DPY30 (Figure 5A) in several MLL1-fusion–mediated leukemia cell lines including MOLM-1345 (MLL1-AF9), THP-146 (MLL1-AF9), KOPN-847 (MLL1-ENL), and RS4;1148 (MLL1-AF4). Cell proliferation was greatly inhibited by DPY30 KD in these cells, as indicated by both the cell growth curves (Figure 5B) and the BrdU incorporation assays (Figure 5C and supplemental Figure 14), although cell cycle arrest was only seen in MOLM-13 and KOPN-8 cells (Figure 5C). No significant effect on apoptosis was seen in these cell lines (Figure 5C). We note that DPY30 KD did not reduce the growth of K562 cells (Figure 3G), leukemia cells driven by tyrosine kinase activation. These results indicate that DPY30 is required for efficient proliferation of the MLL1-fusion–induced leukemia cells. We also evaluated the effect of DPY30 KD on their colony-forming capacity in methylcellulose, which is considered a good indication for self-renewal of leukemia stem cells.49 DPY30 KD abolished the colony formation ability of MOLM-13 cells, resulting in no colonies in the first plating (Figure 5D and supplemental Figure 15A). DPY30 KD also impaired the clonogenicity of both RS4;11 and KOPN-8 cells in serial plating, although the effects in the second plating were less prominent (Figure 5D). We found that the DPY30 KD efficiency was attenuated by the end of the second plating (comparing Figure 5A with supplemental Figure 15B), suggesting that the leukemia cells with low DPY30 level were outcompeted by cells with relatively high DPY30 level in forming colonies in the methylcellulose. These results suggest a detrimental effect of losing DPY30 for the self-renewal ability of the leukemia stem cells contained in at least some of these MLL1-fusion–mediated leukemia cell lines. Toward a mechanistic understanding of the effect of DPY30 KD in these leukemia cells, we confirmed that global H3K4 methylation was reduced by DPY30 KD in MOLM-13 cells (Figure 5E), and it was found that, similar to the effect in HPCs, genes involved in cell proliferation were downregulated by DPY30 KD in MOLM-13 cells (Figure 5F).
DPY30 KD impaired proliferation of MLL1-fusion–mediated leukemia cells. MLL1-fusion–mediated human leukemia cell lines were depleted of DPY30 by shRNAs (#2 and #5 for MOLM-13, and #2 for other cell lines). (A) The DPY30 KD efficiency was determined by qRT-PCR and normalized against ACTB. Averages ± SD from 3 independent biological repeats (infections) are plotted, except for THP-1 cells (2 biological repeats). (B) Cell growth was monitored. Averages ± SD from 3 biological repeats are plotted, except for THP-1 cells (2 biological repeats). (C) Cell proliferation was monitored by BrdU incorporation after culture in the presence of BrdUTP for 1 hour. Percentage of cells in S/G2/M phases were quantified by 7-AAD staining of fixed cells. Apoptotic cells were quantified as Annexin V-positive and 7-AAD negative populations of unfixed cells. Averages ± SD from 3 biological repeats are plotted, except for THP-1 cells (2 biological repeats). *P < .05; ***P < .001 (Student t test). (D) Colony formation assay by serial plating. Averages ± SD from 2 biological repeats are plotted. *P < .05; **P < .01; ***P < .001 (Student t test). (E) Effects of DPY30 KD (short hairpin [sh]#2 and #5) on H3K4 methylation in MOLM-13 cells was examined by western blot analysis. (F) Expression of genes from Figure 1F was also examined for MOLM-13 cells by qRT-PCR against GAPDH. Averages ± SD from 3 biological repeats are plotted. P < .05 (Student t test) between control and either DPY30 shRNAs for all genes except between control and DPY30 sh#5 for MCM3.
DPY30 KD impaired proliferation of MLL1-fusion–mediated leukemia cells. MLL1-fusion–mediated human leukemia cell lines were depleted of DPY30 by shRNAs (#2 and #5 for MOLM-13, and #2 for other cell lines). (A) The DPY30 KD efficiency was determined by qRT-PCR and normalized against ACTB. Averages ± SD from 3 independent biological repeats (infections) are plotted, except for THP-1 cells (2 biological repeats). (B) Cell growth was monitored. Averages ± SD from 3 biological repeats are plotted, except for THP-1 cells (2 biological repeats). (C) Cell proliferation was monitored by BrdU incorporation after culture in the presence of BrdUTP for 1 hour. Percentage of cells in S/G2/M phases were quantified by 7-AAD staining of fixed cells. Apoptotic cells were quantified as Annexin V-positive and 7-AAD negative populations of unfixed cells. Averages ± SD from 3 biological repeats are plotted, except for THP-1 cells (2 biological repeats). *P < .05; ***P < .001 (Student t test). (D) Colony formation assay by serial plating. Averages ± SD from 2 biological repeats are plotted. *P < .05; **P < .01; ***P < .001 (Student t test). (E) Effects of DPY30 KD (short hairpin [sh]#2 and #5) on H3K4 methylation in MOLM-13 cells was examined by western blot analysis. (F) Expression of genes from Figure 1F was also examined for MOLM-13 cells by qRT-PCR against GAPDH. Averages ± SD from 3 biological repeats are plotted. P < .05 (Student t test) between control and either DPY30 shRNAs for all genes except between control and DPY30 sh#5 for MCM3.
Discussion
As the major H3K4 methylation enzymes in mammals, the SET1/MLL complexes are considered potential pharmacologic targets in epigenetic therapeutics. However, a major barrier in targeting these complexes for hematologic diseases is our poor understanding of the role for their H3K4 methylation activity in physiological and pathological hematopoiesis. The direct and important function of Dpy30 for efficient genome-wide H3K4 methylation37 allows the effective functional interrogation of H3K4 methylation in hematopoietic cells by DPY30 depletion. Our work here has shown an important role of DPY30 in the ex vivo proliferation and differentiation of human HPCs as well as zebrafish hematopoiesis. It is possible that either one or more of the SET1/MLL family members are responsible for the H3K4 methylation activity involved in hematopoiesis. As discussed in our previous work37 and elsewhere,50 we cannot formally exclude the possibility that DPY30 may regulate hematopoiesis through functions other than facilitating H3K4 methylation.
Our results strongly suggest a role of DPY30 in direct control of human HPC proliferation through regulation of genes critically involved in DNA replication and cell cycle progression. These findings are consistent with a role of DPY30 in promoting proliferation and preventing senescence in fibroblast cells.51 We have also shown that DPY30 is crucial for the growth of several MLL1-fusion–mediated leukemia cell lines and may be required for the self-renewal capacity of leukemia stem cells in some of those cells. These results suggest that DPY30 and the H3K4 methyltransferase activity of the SET1/MLL complexes may be potential targets for curbing the malignant growth of MLL1-fusion–induced leukemia. Careful gauging on the normal hematopoietic cell activities, however, will be necessary for potential agents targeting DPY30, considering its important roles in normal HPC functions shown here. We note that the effects of DPY30 KD on cell proliferation and global gene expression can be highly dependent on cellular context. Proliferation of HPCs under differentiation conditions (supplemental Figure 6), mouse ESCs,37 and a non-MLL-fusion–driven leukemia cell line (K562 cells) (Figure 3G) is not impaired by DPY30 KD. Furthermore, no significant overrepresentation of major function groups was found for genes significantly affected by DPY30 depletion in mouse ESCs.37 These results suggest that the functionally critical targets of DPY30 and its associated H3K4 methylation are highly cell-type dependent and should be separately characterized in different cells.
The effect of DPY30 KD on CFU-G and CFU-GM formation is consistent with that on the myelomonocytic differentiation in liquid culture (particularly based on the significant effects on the gene expression profiles). The apparently complex phenotypes caused by DPY30 KD in erythroid differentiation probably reflect a differential role of DPY30 at different stages of hematopoietic development and different readouts of the assays. The colony numbers in the colony-forming–cell assays largely reflect the frequency of the functional progenitors in the starting cell population, and the morphologic features of the BFU-E colonies that did form on the DPY30-KD background suggest more efficient erythroid differentiation once they acquire the colony-forming capacity. The differentiation assay in liquid culture, on the other hand, is affected by both the number of the starting progenitors and the differentiation ability of the progenitors. DPY30 KD clearly impaired the proliferation of the erythroid progenitors, and thus resulted in the reduced numbers of differentiated erythroid at the initial stage of erythroid differentiation in the liquid culture. However, DPY30 KD primed the intrinsic transition to erythroid lineage, and, when EPO was present, eventually drove the cells further into more matured erythrocytes producing far more hemoglobin.
In our zebrafish experiments, as dpy30 loss occurred at the early stage of the organism development, the phenotypes could result from the effects on the formation, maintenance, or differentiation of many different cells including HSCs and early HPCs, as well as possible noncell autonomous effects. The severe anemic phenotypes of the dpy30 morphant embryos are shared by embryos targeted for mll52 and for cxxc1 (cfp1, an ortholog for CXXC1 encoding a specific subunit of SET1A/B complexes),53 but these subunits appear to have a different impact on the expression of certain key hematopoietic markers: tal1 and gata1a expression is impaired in the mll,52 but not the cxxc153 morphant embryos; and intriguingly, tal1, gata1a and fli1a signals were all enhanced in the dpy30 morphant embryos compared with the control. A recent screening of chromatin modifiers identified a requirement of many other subunits of SET1 complex in definitive hematopoiesis in zebrafish, primarily based on decreased in situ hybridization signals for c-myb and runx1 in the morphant embryos.54 It is possible that different subunits may differentially impact the hematopoietic marker expression, and different markers may also be differentially affected. Our results in the dpy30 morphant embryos are mostly consistent with a key function of dpy30 in differentiation of HSCs or early HPCs, similar to its demonstrated role in mouse ESCs.37 A more definitive answer to a role of DPY30 in HSC function requires further studies in a hematopoietic-specific Dpy30 knockout mouse model.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank John Parant, Holly Thomas, Susan Farmer, Patty Oden, Peter Jezewski, Henry Jackson, Daniel Gorelick, and Brandie Cline for their tremendous help on our zebrafish experiments; Xinyang Zhao for generously providing K562, HEL, and MOLM-13 cells; Jianjun Chen for THP-1 cells; William Placzek for RS4, 11 cells; Charles Hemenway for KOPN-8 cells; David Langenau for rag1 plasmid; and Xinyang Zhao and William Placzek for the critical reading of the manuscript.
This work was supported by a UAB startup fund (H.J.).
Authorship
Contribution: Z.Y. and J.A. conducted most experiments and analyzed the data; C.C. performed the in situ hybridization experiments; J.H., K.S., and C.W.C. assisted and conducted certain experiments; T.T. coordinated the source of human CD34+ cells and gave valuable suggestions; and H.J. conceived the project, designed experiments, interpreted the results, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hao Jiang, Department of Biochemistry and Molecular Genetics, UAB Stem Cell Institute, University of Alabama at Birmingham, 1825 University Blvd, Birmingham, AL 35294; e-mail: haojiang@uab.edu.
References
Author notes
Z.Y. and J.A. contributed equally to this study.

![Figure 1. DPY30 KD impaired the proliferation of human HPCs. Human CD34+ cells were depleted of DPY30 by 2 different shRNAs (#2 and #5). (A) The KD efficiency was determined by qRT-PCR and normalized against ACTB. Averages ± standard deviation (SD) from 5 independent biological repeats (infections) are plotted. (B) Effect of DPY30 KD (short hairpin [sh]#2 and #5) on global H3K4 methylation level in cells from 2 different sources was determined by western blot analysis. H3K4me1, H3K4me2, and H3K4me3: H3K4 mono-, di-, and tri-methylation, respectively. (C) Various types of myeloid colonies were identified and quantified after the colony-forming–cell assay using control or DPY30-KD cells. Averages ± SD from 3 assays for 1 of 4 independent biological repeats are plotted (the rest are shown in supplemental Figure 2A). P < .01 (Student t test) between the control and either DPY30 shRNAs for all types of colonies. (D) Proliferation of CD34+-gated cells was monitored by BrdU incorporation after culture in the presence of BrdUTP for 12 hours, and the percentage of BrdU-positive cells was quantified from 3 independent biological repeats. *P < .05 (Student t test). (E) Global gene expression was examined by microarray, followed by gene ontology analysis on the genes downregulated by both shRNAs to below three-fourths of the levels in control cells (listed in supplemental Table 3). (F) Expression of selected genes from microarray assays was examined by qRT-PCR against POLR2A. Two different sets of primers were used for MYC. Averages ± SD from 4 independent biological repeats are plotted. P < .05 (Student t test) between control and either DPY30 shRNAs for all genes. (G) Chromatin immunoprecipitation assays for total H3, H3K4me3, and DPY30 binding at the transcription start sites of a negative control gene (OR2J3) and those genes examined in panel F in human CD34+ cells. Averages ± SD from triplicate assays are plotted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/13/10.1182_blood-2014-01-549220/4/m_2025f1.jpeg?Expires=1767793295&Signature=um-vP7wEpwqcBfepwSk8v7kIa3CXRbwuVPGSwvsOJXcbGWqjq6hzGdN1qr8~rX9F3Yloa4zarYQslKEGiUh3QDU0xw77dI9Fg1wJxvQ3xJkNmYycHuQ06bBcqy6G-k1Dv3mizyn-rFqq3q51ISDJFvQkjT0HKkexhdtIqydo5qYQtPjq2TZwPsTl41joDqErZf~AuJzfMYFTFmcphH91VKiVNN7oN964Z6r-UbejJPm~oiexq4bG2~ZIxMTIIh~MUPgey5lF7krMgqgEsdAQjVNbbaGs7TbBiwjUNiwaOGa8yXHOCneGKhWWzXIWr7iABRkF32qBaAQx0UjSekY28A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 2. DPY30 KD impaired myelomonocytic differentiation of human HPCs. Control and DPY30-KD human CD34+ cells were cultured with appropriate cytokine conditions that permit myelomonocytic differentiation. (A) Cell morphology at day 11 under such culture condition. (B) Percentage of myelomonocytic population (CD34−CD11b+) at day 14 in the differentiation assays were quantified. Averages ± SD from 3 biological repeats (infections) are plotted. **P < .01 (Student t test). (C) RNAs from the control and DPY30-KD (by short hairpin [sh]#2) cells on day 9 during myelomonocytic differentiation were subject to microarray assays followed by gene ontology analysis for the genes downregulated over twofold by DPY30 KD (listed in supplemental Table 4). (D) RNAs from the control and DPY30-KD (by sh#2 and #5) cells at day 0 (d0) and day 14 (d14) during myelomonocytic differentiation were subject to quantitative reverse-transcription polymerase chain reaction for selected genes highlighted in supplemental Table 4. Averages ± SD from 2 independent biological repeats are plotted.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/13/10.1182_blood-2014-01-549220/4/m_2025f2.jpeg?Expires=1767793295&Signature=RgDzge1sCk2RqFyJr5BC6T9M5rTS-jcYWHB94lC4GX2Z-S3XuFelozhmq3YLcrvrkG~a~ES6nYQhiL~xQ52EpNvTn5BiWvIWskYwS~XVoMWbLApmTDiYdhfP9kQlMvzMN5~D2uzJT5SJnxMV7WQL2GbOYXJ8E-eI8Uqwg-1h9EkRaWp4QdnWYNfBx688n6uSmru8HratXYcVWluks7exvaHYTsflyvS3kJ5LB4zYPxWHseAFANh2H0drN7AuN7EWwOQKMw-jcfWml8GQzuq23dIO5LyiKuMf6NVc2sG6ltY6mutwIuU7nBRqURFg3KuBWEvaEurYQ6Ccb~BBEvIovw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. DPY30 ortholog is essential for hematopoiesis in zebrafish. (A) Alignment of the human DPY30 and zebrafish dpy30 proteins. Identical amino acids are denoted in bold type and asterisks indicate stop codons. (B) Design of zebrafish dpy30 MOs in relationship to the zebrafish dpy30 gene. The red and gray bars represent the dpy30 MOs. (C and D) Effect of dpy30 MOs on zebrafish blood circulation. (C) Representative images of circulating blood cells (picture frames from the time-lapse video microscopy) in uninjected control zebrafish and zebrafish injected with 0.125× dose of dpy30 MO-1. Green arrows point at the blood cells. (D) Quantification of blood circulation status at indicated time (after fertilization) in uninjected control zebrafish (Control), zebrafish injected with 0.25× dose of dpy30 MO-1 (MO-1), with 0.25× dose of dpy30 MO-1 plus 50 to 100 pg of dpy30 mRNA (MO-1 + mRNA), 1× dose of control MO (cMO), or 0.375× dose of dpy30 MO-2 (MO-2). (E and F) Effect of dpy30 MOs on zebrafish hematopoiesis examined by o-dianisidine staining at 48 hpf. Zebrafish were uninjected (Control), injected with 0.25× dose of dpy30 MO-1 (MO-1), with 0.25× dose of dpy30 MO-1 plus 50 to 100pg of dpy30 mRNA (MO-1 + mRNA), or with 0.375× dose of dpy30 MO-2 (MO-2). (E) Typical results for o-dianisidine staining showing zebrafish with normal red blood cell production and with severe or moderate anemia. Red arrows point at the red blood cells. Note that although some residual red blood cells could sometimes be visualized at the heart region, none was seen in the periphery of the severely anemic fish. These images, from top to bottom, were selected from the Control, MO-1, and MO-1 + mRNA groups, respectively. (F) O-dianisidine staining results were quantified for each category of red blood cell levels (according to the images shown in [E]). Plotted are averages ± SD from 3 independent biological repeats for the left graph (also shown individually in supplemental Figure 10C), and from 2 biological repeats for the right graph (also shown individually in supplemental Figure 11B). **P < .01; ***P < .001 (Student t test). (G) Western blot analysis was used for H3K4 methylation using total extracts from 48 hpf uninjected control zebrafish and zebrafish injected with 0.25× dose of dpy30 MO-1. (H) In situ hybridization for rag1 at day 5 after fertilization in uninjected control zebrafish and zebrafish injected with 0.375× dose of dpy30 MO-2. Dorsal (top) and lateral (bottom) views are shown. The arrow points at the rag1 signal. (I) In situ hybridization for tal1, gata1a, and fli1a at 30 hpf in uninjected control zebrafish and zebrafish injected with 0.25× dose of dpy30 MO-1.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/13/10.1182_blood-2014-01-549220/4/m_2025f4.jpeg?Expires=1767793295&Signature=TWAdS38yEC00WteRDX9hscCSKgv17C4cjAQPd0DHnnWrTjzDbe0VqqUQooQ6BSBl3gGX7PK8S7Eizkz1SEjr19Ii-qoZRej8N~tR99QtYucuoM~3mfJ3hPx6ONR22FjxBu40KXIpqGIWlKthI6Lt8~cHY9mT~lzxf4gNsD4K7yyI0uXfQI4szsN3FwOtfN6AzP-6ARHg6X6PkK0aJtRQbaN~c3mUsv58ylJv1zHUdwMACGpYMgq8J2gmzmz5N7w2V3XDmLWK5K-ZHspiXThcIuVQDbN8t63RzL-lnN5cgZJAbV1GG89WdW4yy7p2oApk07ibG28SIUE16ExnXm4GGQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. DPY30 KD impaired proliferation of MLL1-fusion–mediated leukemia cells. MLL1-fusion–mediated human leukemia cell lines were depleted of DPY30 by shRNAs (#2 and #5 for MOLM-13, and #2 for other cell lines). (A) The DPY30 KD efficiency was determined by qRT-PCR and normalized against ACTB. Averages ± SD from 3 independent biological repeats (infections) are plotted, except for THP-1 cells (2 biological repeats). (B) Cell growth was monitored. Averages ± SD from 3 biological repeats are plotted, except for THP-1 cells (2 biological repeats). (C) Cell proliferation was monitored by BrdU incorporation after culture in the presence of BrdUTP for 1 hour. Percentage of cells in S/G2/M phases were quantified by 7-AAD staining of fixed cells. Apoptotic cells were quantified as Annexin V-positive and 7-AAD negative populations of unfixed cells. Averages ± SD from 3 biological repeats are plotted, except for THP-1 cells (2 biological repeats). *P < .05; ***P < .001 (Student t test). (D) Colony formation assay by serial plating. Averages ± SD from 2 biological repeats are plotted. *P < .05; **P < .01; ***P < .001 (Student t test). (E) Effects of DPY30 KD (short hairpin [sh]#2 and #5) on H3K4 methylation in MOLM-13 cells was examined by western blot analysis. (F) Expression of genes from Figure 1F was also examined for MOLM-13 cells by qRT-PCR against GAPDH. Averages ± SD from 3 biological repeats are plotted. P < .05 (Student t test) between control and either DPY30 shRNAs for all genes except between control and DPY30 sh#5 for MCM3.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/13/10.1182_blood-2014-01-549220/4/m_2025f5.jpeg?Expires=1767793295&Signature=fRncPsY2p6lBWqN94ncth4dJOnoZnpLuyGergHxHqMaiFSD2V2rBfYxutUNPHs1yviCfIpowejcwvgzKx~feMCNZ3DKYKvh0uHnfKIdNYR1ejsm1ZEPSVxlwvIvtvM2jm-yGt7ajNX4vUtUxiR3wAs3wGmdKGDRzm91vEbHNZqn4m9LQZOjNsPy8FMDxuTowQijn1k5ds-Im4ziWrIZ~6vgTIj-~lW4zssCFgfI5NmItqEkM6uc4Wsgt5MAW6OjE1ZCEIXfZ2Vz3YWauiRul-NPc4ybnyGJRCTaeDUGJX02KXfW20mJZDVl2mSjH0WQey184snP6BIz3oKSClKyn-w__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal