Key Points
IRF8K108E mutation causes dendritic cell depletion, defective antigen presentation, and anergic T cells.
IRF8K108E mutant protein is functionally null and shows defective nuclear targeting and increased proteasomal degradation.
Abstract
We have previously reported on a unique patient in whom homozygosity for a mutation at IRF8 (IRF8K108E) causes a severe immunodeficiency. Laboratory evaluation revealed a highly unusual myeloid compartment, remarkable for the complete absence of CD14+ and CD16+ monocytes, absence of CD11c+ conventional dendritic cells (DCs) and CD11c+/CD123+ plasmacytoid DCs, and striking granulocytic hyperplasia. The patient initially presented with severe disseminated mycobacterial and mucocutaneous fungal infections and was ultimately cured by cord blood transplant. Sequencing RNA from the IRF8K108E patient’s primary blood cells prior to transplant shows not only depletion of IRF8-bound and IRF8-regulated transcriptional targets, in keeping with the distorted composition of the myeloid compartment, but also a paucity of transcripts associated with activated CD4+ and CD8+ T lymphocytes. This suggests that T cells reared in the absence of a functional antigen-presenting compartment in IRF8K108E are anergic. Biochemical characterization of the IRF8K108E mutant in vitro shows that loss of the positively charged side chain at K108 causes loss of nuclear localization and loss of transcriptional activity, which is concomitant with decreased protein stability, increased ubiquitination, increased small ubiquitin-like modification, and enhanced proteasomal degradation. These findings provide functional insight into the molecular basis of immunodeficiency associated with loss of IRF8.
Introduction
Primary immunodeficiencies sometimes present with disseminated mycobacterial infection following neonatal vaccination with live Bacillus Calmette-Guérin (BCG).1 In many cases, patients suffer from Mendelian susceptibility to mycobacterial disease, a syndrome caused by infection with weakly virulent mycobacteria such as BCG, with environmental mycobacteria, and/or recurrent infections with virulent mycobacteria (Mycobacteriumtuberculosis) over the life course.2 Approximately half of patients with Mendelian susceptibility to mycobacterial disease have been shown to carry mutations in a small subset of genes involved in interferon (IFN) γ–dependent early immune responses.2,3 Other patients develop disseminated BCG disease as part of a broader pattern of susceptibility to infection (eg, severe combined immunodeficiency).
We have previously reported an infant presenting with severe disseminated BCG infection, oral candidiasis, and severe respiratory viral infection.4 Evaluation of the patient’s blood cellular profile revealed an aberrant myeloid compartment; notably, a complete absence of CD14+ and CD16+ monocytes, absence of CD11c+ conventional dendritic cells (DCs) and CD11c+/CD123+ plasmacytoid DCs, and prominent granulocytic hyperplasia.4 The peripheral blood B-lymphocyte count was also increased in this patient. Production of interleukin (IL) 12 and IFN-γ by blood cells in response to bacterial products or nonspecific stimuli was impaired.4 The patient was cured by cord blood transplant. The phenotype and immunologic features of the patient resembled that of mice bearing partial or complete loss-of-function mutations in Irf8,5-7 including susceptibility to infection with intracellular pathogens such as mycobacteria,7-9 partial (BXH-2; Irf8R294C) or complete (Irf8−/−) absence of CD11c+ CD8α+ DCs and plasmacytoid DCs,5,6,10 and lack of IL-12p40 and IFN-γ production following infection.8,11 Sequencing of IRF8 in this patient identified homozygosity for a unique variant at IRF8 (IRF8K108E). The K108E variant was present in both healthy parents in a heterozygous state and maps to the DNA-binding domain (DBD) of IRF8. This unique patient defines a novel syndrome designated recessive IRF8 DC immunodeficiency.
IRF8 is a member of the IFN regulatory factor (IRF) family and plays essential roles in host defense, hematopoietic differentiation, and immune response, including transcriptional activation in response to IFNs.12 IRF proteins share a highly conserved amino terminal DBD of the helix-turn-helix type (aa1-115) consisting of 5 tryptophan (W) residues that contact DNA at GAAA and AANNNGAAA consensus sequence motifs termed IFN-stimulated response element, found near IFN-regulated genes.13 IRF8 also has an IRF association domain, serving as a recruitment module for other transcription factors, including members of the IRF (eg, IRF1) or E26 transformation-specific (eg, PU.1) families, to activate or repress gene expression,8,14 including genes encoding proteins involved in macrophage antimicrobial functions,15-18 early T helper (Th) 1 polarization of the immune response,19-21 antigen presentation,22 promoting differentiation of myeloid progenitors toward mononuclear lineages, and inducing apoptosis of the granulocytic lineage.23 Finally, IRF8 is a key regulator of pathological inflammation: inactivation of Irf8 protects against neuroinflammation in Plasmodium berghei–infected mice,24 and association studies have identified IRF8-associated polymorphisms as a key genetic risk for low monocyte counts,25 rheumatoid arthritis,26 systemic lupus erythematosus,27 ulcerative colitis,28 Crohn's disease,29,30 and multiple sclerosis31 in humans. Hence, characterizing the genes that are bound and regulated by IRF8 in human cells may shed important light on proteins and pathways that underlie pathological inflammation in humans.
The K108E patient is the only known example of recessive IRF8 deficiency in humans. Comparative studies in primary cells from this individual offer a unique window of opportunity to identify and characterize the transcriptional targets of IRF8 in vivo and in the context of its normal physiological function in human cells. These studies may identify proteins and pathways that are required for ontogeny and maturation of myelomonocytic cells including effective antimicrobial defenses, as well as pathways activated during pathological inflammation. Finally, biochemical characterization of the mechanistic basis of loss of function in K108E may provide valuable information of pathogenesis of DC deficiency in this patient.
Materials and methods
Plasmid construction
A 500-bp portion of the Isg15 gene promoter region containing an IFN-stimulated response element was polymerase chain reaction amplified from genomic DNA using oligonucleotide primers 5′-ACTGACTCGAGTGC TGGGATCAAAGGTGTGC-3′and 5′-GTCAGAAGCTTGAGTCTGCTTCTGGCTGCTT-3′ and cloned into the luciferase reporter vector pGL3 (Promega, Madison, WI) using XhoI and HindIII restriction sites. The IL-12p40-pGL3, IRF8-hemagglutinin (HA)-pcDNA3, and IRF1-pcDNA3 vectors were generated as previously described.32 The V5-small ubiquitin-like modifier (SUMO)3 and HA-sentrin specific peptidase (SENP)1 plasmids were as previously described33 and were kindly provided by Keiko Ozato (National Institutes of Health).
Cell culture, transfections, luciferase assays, and western blots
RAW 264.7 and HEK 293T cells were maintained in Dulbecco’s modified Eagle medium (Wisent) supplemented with 10% heat-inactivated fetal bovine serum (Wisent). For luciferase assays, RAW cells (1 × 105 cells per well) were transfected using Lipofectamine PLUS (Invitrogen Life Technologies) with 400 ng luciferase pGL3 reporter, 100 ng Renilla internal control, and variable amounts of pcDNA3 expression vectors. Luciferase activity was assayed 24 hours following transfection using a dual luciferase assay system (Promega). Whole cell extracts were analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and incubated with mouse anti-HA monoclonal antibody (mAb) (1:100; Covance Research Products, Princeton, NJ) followed by washing and incubation with an anti-mouse secondary antibody conjugated to horseradish peroxidase (1:20 000; GE Healthcare).
Immunoprecipitation, ubiquitination, and SUMOylation assays
HEK 293T cells were transfected and washed with cold phosphate-buffered saline (PBS) and lysed for 30 minutes on ice in lysis buffer (50 mM tris[hydroxymethyl]aminomethane HCl pH 7.5, 150 mM NaCl, 1% Triton X-100, 0.1% SDS) supplemented with protease inhibitors. Lysates (500 µg) were incubated with 2 µg anti-IRF8 antibody C-19 (Santa Cruz Biotechnology Inc., Santa Cruz, CA) overnight at 4°C. Supernatants were collected and incubated with 50 µL of recombinant Protein G Agarose beads at 4°C for 4 hours. Immune complexes were resuspended in 50 µL Laemmli sample buffer (60 mM tris[hydroxymethyl]aminomethane HCl pH 6.8, 2% SDS, 10% glycerol, 5% b-mercaptoethanol, 0.02% bromophenol blue) and heated at 95°C for 5 minutes. Samples were subjected to SDS-PAGE and immunoblotted either with anti-Ub P4D1 (1:250; Santa Cruz Biotechnology Inc.), anti-IRF8 C-19 (1:250; Santa Cruz Biotechnology Inc.), or anti-V5 (1:5000; Invitrogen Life Technologies).
Immunofluorescence
Stably transfected RAW macrophage clones were seeded onto glass coverslips (1 × 105 cells per well). Cells were washed with PBS 24 hours later and fixed in 4% paraformaldehyde in PBS for 15 minutes, and then permeabilized with 0.1% Triton X-100 for 15 minutes. Coverslips were blocked with 0.1% bovine serum albumin (BSA) in PBS for 1 hour, then incubated with mouse anti-HA mAb (1:200) for 1 hour, and washed with 0.1% BSA in PBS. The coverslips were then incubated for 1 hour with goat anti-mouse Cy3-conjugated secondary antibody (1:100) and washed with 0.1% BSA in PBS and then with 4′,6-diamidino-2-phenylindole (DAPI) in PBS. Coverslips were mounted on microscope slides using Permafluor Aqueous Mounting Medium (Thermo Scientific). Confocal microscopy was conducted with a Zeiss LSM5Pascal laser scanning confocal microscope. All image analyses were performed using LSM5 Image.
Cellular fractionation
Stably transfected RAW macrophage clones were treated or not with IFN-γ (400 U/mL; 3 hours, 37°C). Cell were washed with PBS, resuspended in buffer A (10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA), and incubated on ice for 15 minutes. NP-40 (Sigma-Aldrich) was added at a final concentration of 0.6%, vortexed for 10 seconds, and centrifuged for 1 minute (13 000g, 4°C). The supernatant containing the cytoplasmic fraction was collected. The pellet was then washed twice with PBS and resuspended in buffer B (20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid pH 7.9, 25% glycerol, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA) and incubated at 4°C on a rotator for 40 minutes. Samples were centrifuged for 5 minutes (13 000g, 4°C), and the supernatant containing the nuclear fraction was collected. Both buffers were supplemented with 2 mM dithiothreitol and protease inhibitors. Fractions were separated by SDS-PAGE and immunoblotted with anti-IRF8 C-19, anti-RNA Pol II sc-899, and anti-GAPDH 6C5 antibodies from Santa Cruz Biotechnology.
RNA sequencing (RNA-seq)
The McGill Institutional Review Board approved the use of archival frozen material (peripheral blood mononuclear cells [PBMCs]) for RNA extraction and RNA-seq. Total PBMCs RNA was extracted using RNeasy columns (Qiagen), and integrity was assessed on a Bioanalyzer RNA pico chip. The RNA-seq libraries were sequenced on a HiSequation 2000 sequencer. Using the Trimmomatic software,34 raw read sequences were trimmed for minimum quality at the 3′ end (phred score of at least 30), cleaned of adapter traces, and filtered for a final minimum length of 32 bp. Alignment to the University of California, Santa Cruz hg19 reference genome (Illumina iGenomes) sequence was performed using TopHat2,35 and gene expression was quantified by counting the number of uniquely mapped reads with HTSeq (http://www-huber.embl.de/users/anders/HTSeq/). Normalization and differential expression analysis between patient and controls was conducted using the edgeR Bioconductor package.36 We retained genes that had an expression level of at least 1 count per million reads (CPM) in at least 1 of the 6 samples. The K108E sample was first compared with all control samples; we also compared the Ctl1 sample with all other control samples. Differentially expressed transcripts were identified as showing a more than threefold difference between the K108E and Ctl1 sample (P value ≤.05 for K108E vs all Ctls), while correcting for minor variations among all control samples (P value ≥.05). Data are shown as heatmap (Log2 CPM) created using MeV.37 Gene ontology (GO) enrichment analysis was performed using the DAVID Web site.38 The differential gene expression profile was also subjected to gene set enrichment analysis (GSEA) using GSEA39 with MSigDB public immunologic gene signatures. Among the enriched gene signatures, 8 representative signatures were selected for graphical representation (GSE22886_neutrophil_vs_monocyte_up, GSE22886_neutrophil_vs_monocyte_dn, GSE22886_neutrophil_vs_DC_dn, GSE9988_lipopolysaccharide (LPS)_vs_vehicule_treated_up, GSE22886_Ctrl_vs_LPS_24h_DC_dn, GSE22886_naive_CD4_Tcell_vs_48h_act_Th1_up, Goldrath_naive_vs_Eff_CD8_Tcell_dn, GSE11057_naive_vs_memory_CD4_Tcell_dn, GSE13411_plasma_cell_vs_memory_Bcell_up, GSE22668_Naive_Bcell_vs_BM_Plasma_cell_UP).
Results
Hematologic profile of the IRF8K108E patient
A 3-month-old infant patient homozygous for a loss-of-function mutation at IRF8 (K108E) presented with symptoms of immunodeficiency, which included disseminated BCG infection (following neonatal BCG vaccination), oral candidiasis, and severe respiratory viral infection. Examination of blood profile and fluorescence-activated cell sorter–assisted immunophenoptyping (Figure 1), showed persistent and extreme neutrophilia (note log scale), absolute monocytopenia (CD14+ and CD16+ cells), anemia, and thrombocytopenia. The patient was not lymphopenic but rather developed a marked B lymphocytosis (note log scale), and to a lesser extent NK lymphocytosis, as her clinical status evolved toward an increasingly inflammatory picture (Figure 1). This B lymphocytosis was accompanied by elevated plasma immunoglobulins (supplemental Figure 1; see the Blood Web site) with strikingly high levels of IgM (1.2-5.5 g/L). This was treated as hemophagocytic lymphohistiocytosis using a modified version of the HLH-2004 protocol (Figure 1, blue triangle) resulting in decline in neutrophils and B and NK cell counts, accompanied by clinical stabilization. The patient moved rapidly into conditioned hematopoietic stem cell transplant (Figure 1, red triangle) at 9 months of age, following which all hematologic parameters were ultimately normalized, including appearance of circulating monocytes. Immunologic reconstitution was fully achieved including normal humoral responses to vaccines.
Blood cellular profile of patient K108E prior to and post stem cell transplant. Hematologic profiles were obtained by standard automated measurements (neutrophils, platelets, hemoglobin), and immune cell populations (total T lymphocytes, CD4+ T cells, CD8+ T cells, natural killer [NK] cells, B lymphocytes, monocytes) were established by fluorescence-activated cell sorter using cell-specific markers. (The degree of monocytopenia was inconsistently evident from machine differential counts beyond the patient’s initial presentation; those derived from flow cytometric evaluation are shown.) The patient was treated as hemophagocytic lymphohistiocytosis using a modified version of the HLH-2004 protocol (blue triangle) with resulting decline in neutrophil, B, and NK cell counts, accompanied by clinical stabilization. The patient then moved rapidly into conditioned hematopoietic stem cell transplant (red triangle; 9 months of age).
Blood cellular profile of patient K108E prior to and post stem cell transplant. Hematologic profiles were obtained by standard automated measurements (neutrophils, platelets, hemoglobin), and immune cell populations (total T lymphocytes, CD4+ T cells, CD8+ T cells, natural killer [NK] cells, B lymphocytes, monocytes) were established by fluorescence-activated cell sorter using cell-specific markers. (The degree of monocytopenia was inconsistently evident from machine differential counts beyond the patient’s initial presentation; those derived from flow cytometric evaluation are shown.) The patient was treated as hemophagocytic lymphohistiocytosis using a modified version of the HLH-2004 protocol (blue triangle) with resulting decline in neutrophil, B, and NK cell counts, accompanied by clinical stabilization. The patient then moved rapidly into conditioned hematopoietic stem cell transplant (red triangle; 9 months of age).
Expression studies in blood cells from the IRF8K108E patient identify defects in antigen-presenting cells (APCs) and in T-cell activation
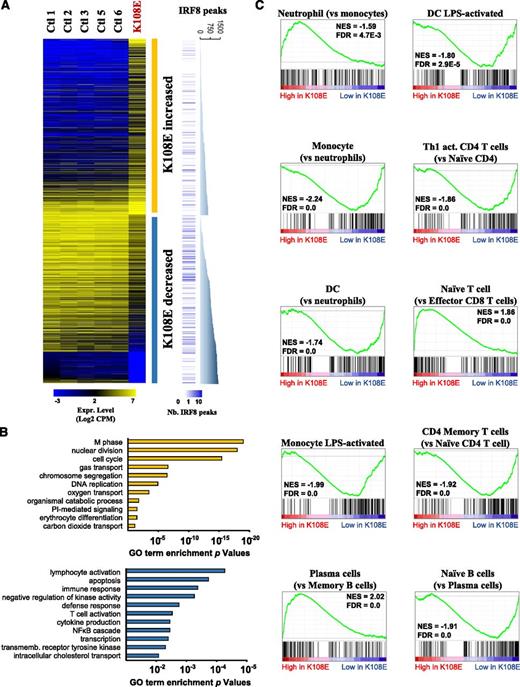
To gain insight into functional consequences of loss of IRF8 and to identify IRF8 transcriptional targets in primary human cells in vivo, we used RNA-seq to compare expression profiles of PBMCs from 5 healthy controls with that of the K108E patient obtained prior to curative cord blood transplant. This analysis (supplemental Table 1) detected 724 genes with increased expression and 694 genes with decreased expression in K108E PBMCs compared with controls (Figure 2A). A priori, these lists are expected to contain the following: (a) genes that are directly bound and transcriptionally regulated by IRF8 (and whose expression is lost in the patient); and (b) genes that are differentially expressed among cell types that are more or less numerous in patient vs controls, the ontogeny and numbers of which are regulated by IRF8. GO annotations (Figure 2B) indicated that transcripts elevated in the K108E patient were associated predominantly with cell cycle and cell division, in agreement with the increased number of CD34+ hematopoietic progenitors, and with the myeloproliferative profile (granulocytic hyperplasia) of the patient.4 On the other hand, transcripts linked to innate immunity were relatively depleted in the K108E patient, in agreement with the patient’s immunodeficiency phenotype and lack of monocytes and DCs.
IRF8K108E mutation modifies global gene expression profile of blood cells. (A) RNA extracted from PBMCs of 5 healthy individuals and from the IRF8K108E patient were sequenced (RNA-seq). The expression level of significantly dysregulated genes in the K108E sample, shown as Log2 of sequence read CPM, are presented in a heatmap together with the expression level of the control samples. There are 724 genes that exhibit increased and 694 genes that have decreased expression in the K108E PBMCs sample. The number of IRF8 binding sites found in the vicinity of each dysregulated gene is represented by blue bars. For the 2 groups of genes, a graph shows the cumulative number of IRF8 binding sites found in proximity of these genes in chromatin from activated macrophages. (B) GO enrichment analysis of genes that show either increased (yellow) or decreased (blue) expression in K108E compared with controls; the degree of statistical significance is shown. (C) The K108E expression profile was subjected to GSEA39 to identify specific immune cell signatures either overrepresented or depleted in K108E compared with controls. Immune cell signatures were produced by pairwise comparisons of public data sets (see “Materials and methods”) and are indicated on top of each graph. GSEA graphs illustrate the cumulative enrichment score in each specific gene signature comparison; the occurrence of the cell signature genes is identified as individual black lines over the distribution of the K108E patient gene profile. Normalized enrichment scores (NES) and false-discovery rate (FDR) are shown for each displayed analysis.
IRF8K108E mutation modifies global gene expression profile of blood cells. (A) RNA extracted from PBMCs of 5 healthy individuals and from the IRF8K108E patient were sequenced (RNA-seq). The expression level of significantly dysregulated genes in the K108E sample, shown as Log2 of sequence read CPM, are presented in a heatmap together with the expression level of the control samples. There are 724 genes that exhibit increased and 694 genes that have decreased expression in the K108E PBMCs sample. The number of IRF8 binding sites found in the vicinity of each dysregulated gene is represented by blue bars. For the 2 groups of genes, a graph shows the cumulative number of IRF8 binding sites found in proximity of these genes in chromatin from activated macrophages. (B) GO enrichment analysis of genes that show either increased (yellow) or decreased (blue) expression in K108E compared with controls; the degree of statistical significance is shown. (C) The K108E expression profile was subjected to GSEA39 to identify specific immune cell signatures either overrepresented or depleted in K108E compared with controls. Immune cell signatures were produced by pairwise comparisons of public data sets (see “Materials and methods”) and are indicated on top of each graph. GSEA graphs illustrate the cumulative enrichment score in each specific gene signature comparison; the occurrence of the cell signature genes is identified as individual black lines over the distribution of the K108E patient gene profile. Normalized enrichment scores (NES) and false-discovery rate (FDR) are shown for each displayed analysis.
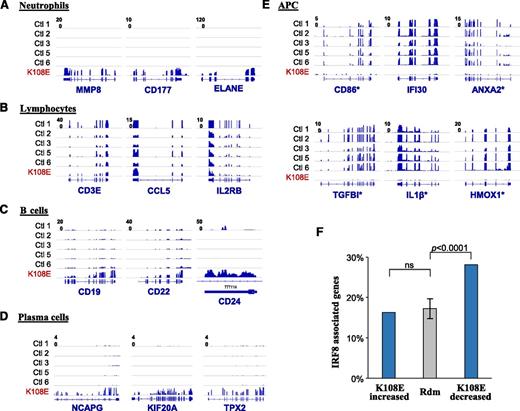
To study the effect of the IRF8K108E variant on ontogeny and activity of individual blood cell populations, we compared the K108E-associated gene expression profile with discrete immunologic cell-specific transcript signatures obtained by GSEA.39 As expected from the extreme neutrophilia seen in the K108E patient, the GSEA analysis shows strong enrichment of neutrophil-specific signatures exemplified by neutrophil-specific transcripts MMP8, CD177, and ELANE (Figures 2C and 3A). Moreover, and in agreement with the B-cell lymphocytosis noted in the patient, a B-cell–specific signature (eg, CD19, CD22, CD24) was overrepresented (Figures 2C and 3C); more specifically, a plasma cell signature (eg, NCAPG, KIF20A, and TPX2) was enriched compared with naïve B or memory B cells (Figure 3D), perhaps in keeping with elevated plasma immunoglobulin in the patient (supplemental Figure 1). A close examination the RNA-seq data over the immunoglobulin heavy chain locus also confirmed prominent IgM expression (data not shown). However, most striking was a decrease in monocyte- and DC-specific signatures, and in LPS-responsive transcripts in the patient (Figure 2C), with CD86, IFI30, ANXA2, TGFBI, IL-1B, and HMOX1 providing the clearest examples of this myeloid signature (Figure 3E). This is in agreement with the known emptiness of the antigen-presenting compartment in this patient (Figure 1). Surprisingly, this analysis also clearly showed that despite being in the normal range (Figures 1 and 3B), the patient’s T cells are in a more naïve state, with a striking depletion of Th1 activated CD4+ T cell, and of CD4+ and of CD8+ antigen-experienced memory T-cell gene signatures (Figure 2C). This suggests that the primary defect of APCs in K108E is associated with a secondary defect in lymphoid cells.
IRF8 directly controls gene expression in APCs. The RNA-seq data were queried for hematopoietic cell–specific genes, and the density of sequence reads mapped to individual gene exons is shown for 5 controls and for K108E. (A) Neutrophil genes are enriched in K108E, and T lymphocyte genes (B) are similar in controls and patient K108E. B lymphocyte (C) and plasma cell (D) specific genes are enriched in patient K108E. (E) Transcripts expressed in APCs are depleted in K108E in agreement with published cellular immunophenotyping data of the K108E patient.4 The asterisk identifies genes that were found to be direct IRF8 targets in chromatin immunoprecipitation sequencing (ChIP-seq) experiments. (F) The list of genes differentially expressed in K108E was compared with a list of direct IRF8 targets (containing an IRF8 binding site within 20 kb from the transcription start site) and obtained by ChIP-seq that we recently published.24 Genes bearing IRF8 binding sites are significantly enriched in the list of genes depleted in K108E, compared with genes of increased expression in K108E, or to a set of 10 randomly generated gene sets. The P value is calculated using Fisher’s exact test.
IRF8 directly controls gene expression in APCs. The RNA-seq data were queried for hematopoietic cell–specific genes, and the density of sequence reads mapped to individual gene exons is shown for 5 controls and for K108E. (A) Neutrophil genes are enriched in K108E, and T lymphocyte genes (B) are similar in controls and patient K108E. B lymphocyte (C) and plasma cell (D) specific genes are enriched in patient K108E. (E) Transcripts expressed in APCs are depleted in K108E in agreement with published cellular immunophenotyping data of the K108E patient.4 The asterisk identifies genes that were found to be direct IRF8 targets in chromatin immunoprecipitation sequencing (ChIP-seq) experiments. (F) The list of genes differentially expressed in K108E was compared with a list of direct IRF8 targets (containing an IRF8 binding site within 20 kb from the transcription start site) and obtained by ChIP-seq that we recently published.24 Genes bearing IRF8 binding sites are significantly enriched in the list of genes depleted in K108E, compared with genes of increased expression in K108E, or to a set of 10 randomly generated gene sets. The P value is calculated using Fisher’s exact test.
We also compared the list of transcripts altered in the K108E patient (supplemental Table 1) with a list of direct transcriptional targets of IRF8 that we identified by sequencing of IRF8-specific chromatin (ChIP-seq) immunoprecipitated from a macrophage cell line treated with IFN-γ.24 We noted a strong and significant depletion of transcripts from genes close to IRF8 binding sites in the K108E patient (Figures 2A and 3F). Such genes represent major and direct IRF8 transcriptional targets in vivo in myeloid cells, expression of which is abrogated in the patient. This list defines the true IRF8 myeloid transcriptome (supplemental Table 2) and includes proinflammatory molecules (IL-1β, C1QA, C2, CCL3, CCL4) and receptors (CCR5, CCR7, CSF1R, TGFBR2), as well as cell surface proteins that define myeloid (CD14, CD163, CD300e) cells. These results show that in humans, loss of IRF8 activity is associated with depletion of myelomonocytic cells, impaired T-cell function, in addition to loss of IRF8-dependent transcription in these and other blood cells.
Impaired nuclear targeting, decreased stability, and altered posttranslational modification of the IRF8K108E variant
IRF8 can repress Isg15 gene expression, acting to inhibit IRF1-dependent activation of certain genes, including Isg15.40 Although wild-type (WT) IRF8 induces dose-dependent repression of the Isg15 promoter activity, this activity is lost in the E108 mutant (Figure 4A). IRF8 also inhibits IRF1-dependent transcriptional activation of the Isg15 reporter, and this inhibitory effect is lost in E108 (Figure 4B). Conversely, and as opposed to K108, the E108 variant fails to promote IRF1-dependent transactivation of an IL-12p40 luciferase reporter construct (Figure 4C).4 To gain insight into the structural requirements at K108 that are required for function, we tested other mutants at position 108. The Q108 variant, in which the positively charged side chain of lysine is eliminated (uncharged glutamine), cannot transactivate the IL-12p40 reporter, whereas an R108 variant that maintains a positively charged side chain of similar size retains transcriptional activity (Figure 4C). Finally, H108, which harbors an imidazole ring and a partial positive charge, shows significant transcriptional activity (Figure 4C). Immunoblotting experiments show that all variants are expressed at similar levels in transfected cells. Also, the R108 mutant shows electrophoretic mobility (a function of charge and folding) similar to K108, but distinct from E108 (Figure 4D). K108 maps to a short β-strand making a hydrogen bond (3.3 Å) and docking IRF8 onto DNA; mutation to E108 creates repulsion, allowing for a water molecule to fill the space and to destabilize the binding.3 Molecular modeling of the additional mutants reveals that H108 has a protonable side chain (pK ∼7) that can act as a donor for hydrogen bonding depending on the environment (supplemental Figure 2). This is consistent with the partial loss of transactivation seen in this mutant. Finally, R108 is structurally similar to K108 and is fully functional (supplemental Figure 2). These results indicate that it is the loss of positive charge, rather than the gain of a negative charge, that is responsible for the loss of transcriptional function of E108.
Loss of transcriptional function in the IRF8K108E mutant. (A) RAW macrophages were transiently transfected with increasing amounts of WT (K108) or IRF8K108E variant (E108) along with a firefly luciferase reporter construct containing the Isg15 promoter and a Renilla luciferase internal control, and luciferase activity was assayed 24 hours later. (B) RAW cells were transiently transfected as in panel A with the addition of IRF1. (C) RAW cells were transiently transfected with expression vectors coding for K108 or with the E108, R108, Q108, and H108 mutants, along with or without IRF1 and an IL-12p40-pGL3 luciferase reporter construct and a Renilla luciferase internal control. Results are represented as means ± standard deviation (SD) of 3 independent experiments. (D) Immunoblots showing similar expression level of K108, and of the E108, R108, Q108, and H108 mutants in transfected cells, and using a mouse anti-HA mAb. Actin expression was evaluated as a loading control.
Loss of transcriptional function in the IRF8K108E mutant. (A) RAW macrophages were transiently transfected with increasing amounts of WT (K108) or IRF8K108E variant (E108) along with a firefly luciferase reporter construct containing the Isg15 promoter and a Renilla luciferase internal control, and luciferase activity was assayed 24 hours later. (B) RAW cells were transiently transfected as in panel A with the addition of IRF1. (C) RAW cells were transiently transfected with expression vectors coding for K108 or with the E108, R108, Q108, and H108 mutants, along with or without IRF1 and an IL-12p40-pGL3 luciferase reporter construct and a Renilla luciferase internal control. Results are represented as means ± standard deviation (SD) of 3 independent experiments. (D) Immunoblots showing similar expression level of K108, and of the E108, R108, Q108, and H108 mutants in transfected cells, and using a mouse anti-HA mAb. Actin expression was evaluated as a loading control.
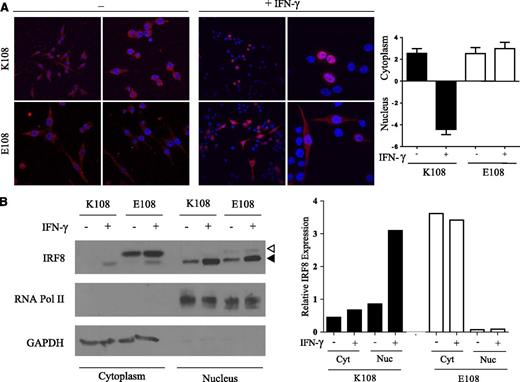
At steady state, IRF8 is expressed at low levels in the cytoplasm. In response to exposure to microbial products or to IFN-γ, IRF8 expression is induced and IRF8 is recruited to the nucleus.20 We generated macrophages stably expressing HA-tagged K108 or variants at this position and monitored protein expression and subcellular localization by immunofluorescence, and by biochemical cell fractionation. In resting macrophages, all variants are found in the cytoplasm (Figure 5A; supplemental Figure 3). Following stimulation with IFN-γ for 3 hours, K108 and the structurally similar R108 are both translocated to the nucleus where they colocalize with DAPI staining, whereas H108 shows reduced nuclear translocation. This IFN-γ–induced nuclear translocation is lost in both the E108 and Q108 variants, which are mainly retained in the cytoplasm (Figure 5A; supplemental Figure 3). Cytoplasmic retention of E108 was further confirmed by cell fractionation experiments (Figure 5B). In these experiments, we used an anti-IRF8 antibody that distinguishes the slower-migrating transfected E108 mutant from the faster-migrating endogenous K108 protein. These experiments also allowed us to monitor a possible effect of the mutant variant on the localization of WT protein when expressed in the same cell. Following IFN-γ stimulation, E108 is predominantly retained in the cytoplasm and barely detected in nuclear fractions, whereas K108 is efficiently translocated into the nucleus. Expression of the E108 mutant does not affect localization or nuclear translocation of endogenous K108 present in RAW cells (Figure 5B). Loss of nuclear translocation likely contributes to the abrogated transcriptional activity of the E108 and Q108 variants (Figure 4) by preventing access of these proteins to DNA.
Defective nuclear translocation of the IRF8K108E mutant. (A) RAW macrophages stably expressing HA-tagged K108 and E108 variants were stimulated or not with IFN-γ (400 U/mL) for 3 hours. Cells were fixed, permeabilized, and incubated with mouse anti-HA mAb followed by mouse Cy3-conjugated secondary antibody (red) and then with DAPI to stain nuclei (blue). Cellular localization was analyzed by confocal microscopy using a Zeiss LSM5 Pascal laser scanning confocal microscope. All image analyses were performed using LSM5 Image. Two different images of each condition are shown. Images are representative of 3 independent experiments. Mean fluorescence intensity of Irf8 (red) staining was measured using Image J. Ratios of cytoplasmic over nuclear fluorescence are represented as means ± SD of 10 independent measurements. (B) RAW macrophages stably expressing HA-tagged K108, E108 were stimulated or not with IFN-γ (400 U/mL) for 3 hours. Cell lysates were subjected to cellular fractionation to separate the cytoplasmic and nuclear fractions and immunoblotted with anti-IRF8, anti-RNA Pol II (nuclear control), and anti-GAPDH (cytoplasmic control) antibodies. The white triangle represents transfected E108, and the black triangle represents endogenous K108. The relative nuclear and cytoplasmic distribution of the total K108 and E108 proteins was evaluated and is shown.
Defective nuclear translocation of the IRF8K108E mutant. (A) RAW macrophages stably expressing HA-tagged K108 and E108 variants were stimulated or not with IFN-γ (400 U/mL) for 3 hours. Cells were fixed, permeabilized, and incubated with mouse anti-HA mAb followed by mouse Cy3-conjugated secondary antibody (red) and then with DAPI to stain nuclei (blue). Cellular localization was analyzed by confocal microscopy using a Zeiss LSM5 Pascal laser scanning confocal microscope. All image analyses were performed using LSM5 Image. Two different images of each condition are shown. Images are representative of 3 independent experiments. Mean fluorescence intensity of Irf8 (red) staining was measured using Image J. Ratios of cytoplasmic over nuclear fluorescence are represented as means ± SD of 10 independent measurements. (B) RAW macrophages stably expressing HA-tagged K108, E108 were stimulated or not with IFN-γ (400 U/mL) for 3 hours. Cell lysates were subjected to cellular fractionation to separate the cytoplasmic and nuclear fractions and immunoblotted with anti-IRF8, anti-RNA Pol II (nuclear control), and anti-GAPDH (cytoplasmic control) antibodies. The white triangle represents transfected E108, and the black triangle represents endogenous K108. The relative nuclear and cytoplasmic distribution of the total K108 and E108 proteins was evaluated and is shown.
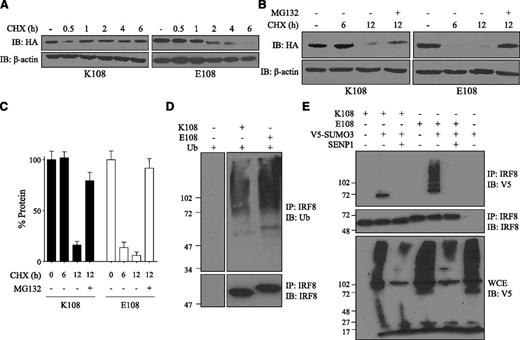
Reduced transactivation function and defective nuclear translocation as well as altered electrophoretic mobility suggested that the K108E mutation may also affect protein folding, with pleiotropic consequences on stability, subcellular targeting, and overall function. Stability of the K108 and E108 variants was investigated in cells stably expressing each protein and following treatment with the protein synthesis inhibitor cycloheximide (CHX). Levels of each protein were monitored over time in CHX-treated cells by immunoblotting (Figure 6A). Over a 6-hour period, K108 protein was stable in CHX-treated cells, suggesting a half-life of more than 6 to 8 hours; under the same conditions, there was a rapid disappearance of E108 variant, with <10% protein remaining at 6 hours (Figure 6A). Enhanced degradation of E108 was proteasome dependent and sensitive to the proteasome inhibitor MG132 (Figure 6B-C). The results suggest reduced stability and increased degradation of the E108 variant.
Decreased stability of the IRF8K108E mutant. (A) RAW macrophages stably expressing HA-tagged K108 or E108 variant were treated with CHX for the indicated periods of time (hours) and for up to 6 hours. Cell lysates were subjected to SDS-PAGE and immunoblotted with a mouse anti-HA mAb. Actin was used as an internal loading control. (B) RAW macrophages stably expressing HA-tagged K108 or E108 were treated with CHX for 6 hours and 12 hours with or without the proteasome inhibitor MG132 (10 µM), and cell lysates were analyzed by immunoblotting. (C) Relative protein expression was assessed by densitometry using ImageJ. Results are represented as means ± SD of 3 independent experiments. (D) HEK 293T cells were transiently transfected with K108 or E108 variant, along with ubiquitin, and 24 hours later, cells were treated with MG132. IRF8 was immunoprecipitated from cell lysates followed by immunoblotting with anti-ubiquitin antibody. IRF8 was also detected by immunoblotting from total cell lysates. The left and right panels come from the same experiment, membrane and film, and thus can be used for comparison. (E) HEK 293T cells were transiently transfected with K108 or E108 variant, with or without V5-tagged SUMO3 and/or HA-tagged SENP1. Cell lysates were immunoprecipitated with anti-IRF8 antibody and immunoblotted with anti-V5 antibody. Total cell extracts were also immunoblotted with anti-V5 antibody to show SUMOylation of other proteins and SENP1 de-SUMOylating activity.
Decreased stability of the IRF8K108E mutant. (A) RAW macrophages stably expressing HA-tagged K108 or E108 variant were treated with CHX for the indicated periods of time (hours) and for up to 6 hours. Cell lysates were subjected to SDS-PAGE and immunoblotted with a mouse anti-HA mAb. Actin was used as an internal loading control. (B) RAW macrophages stably expressing HA-tagged K108 or E108 were treated with CHX for 6 hours and 12 hours with or without the proteasome inhibitor MG132 (10 µM), and cell lysates were analyzed by immunoblotting. (C) Relative protein expression was assessed by densitometry using ImageJ. Results are represented as means ± SD of 3 independent experiments. (D) HEK 293T cells were transiently transfected with K108 or E108 variant, along with ubiquitin, and 24 hours later, cells were treated with MG132. IRF8 was immunoprecipitated from cell lysates followed by immunoblotting with anti-ubiquitin antibody. IRF8 was also detected by immunoblotting from total cell lysates. The left and right panels come from the same experiment, membrane and film, and thus can be used for comparison. (E) HEK 293T cells were transiently transfected with K108 or E108 variant, with or without V5-tagged SUMO3 and/or HA-tagged SENP1. Cell lysates were immunoprecipitated with anti-IRF8 antibody and immunoblotted with anti-V5 antibody. Total cell extracts were also immunoblotted with anti-V5 antibody to show SUMOylation of other proteins and SENP1 de-SUMOylating activity.
IRF8 is targeted for proteasome degradation following ubiquitination by the E3 ubiquitin ligase Cbl.41 In addition, SUMOylation via SUMO2 and SUMO3 keeps IRF8 in a repressed functional state and can also target IRF8 for proteasomal degradation.33 Following activation of macrophages, IRF8 is deSUMOylated by SENP1, which releases IRF8 for transcriptional activity.33 We investigated the effect of K108E on regulatory posttranslational modification of IRF8 (Figure 6D-E). In transfected HEK 293T cells, we observed for both WT and K108E mutant, a high-molecular-mass smear typical of ubiquitinated proteins, but this smear was more intense for E108, suggesting increased ubiquitination of the mutant compared with WT (Figure 6D). To test the effect of K108E on the state of IRF8 SUMOylation, HEK 293T cells were transfected with either K108 or with E108, and with V5-tagged SUMO3, with or without the de-SUMOylation enzyme SENP1. SUMO3-dependent and SENP1-sensitive SUMOylation of K108 was detected in transiently transfected HEK293T cells. However, for the E108 variant, there was a strong increase in SUMO3-dependent and SENP1-sensitive conjugation compared with K108, with appearance of higher-molecular-weight forms unique to the E108 variant, with all forms being sensitive to SENP1 (Figure 6E). Together, these results indicate increased ubiquitination and increased SUMOylation of the K108E variant, which is concomitant to decreased protein stability and increased proteasome-dependent degradation.
Discussion
Patient IRF8K108E is the only reported case of recessive IRF8 deficiency in humans to date. This was characterized by complete absence of circulating blood DCs and monocytes in an overall myeloproliferative picture, together with striking susceptibility to infection including mycobacterial disease. This unique patient offers the rare occasion to study the role of monocytes and DCs in vivo, and to characterize possible pleiotropic effects of their absence on the activity of other cell types of the immune system. Critically, comparative transcript profiling studies in IRF8-deficient primary human cells from this patient offer the exceptional opportunity to identify IRF8 transcriptional targets of relevance in vivo.
Although the patient was cured by cord blood transplant, we were able to analyze 1 archival sample of PBMCs, cryopreserved prior to transplant. RNA-seq and gene profiling studies showed that, as expected, IRF8K108E was associated with depletion of transcripts associated with mononuclear phagocytes, whereas it was enriched for transcripts from the granulocytic lineage, in agreement with the immunologic profile. A novel finding was the depletion of gene signatures associated with T-lymphocyte activation in the patient, and this for both the CD4+ and the CD8+ lineages, which rather displayed signatures characteristic of naïve T cells. This occurred despite normal numbers of circulating T cells in the patient, and despite intense antigenic exposure taking place during the multiple episodes of infection (bacterial, viral, and fungal) in this patient. These results extend in vitro studies with cultured T cells from the patient showing poor responsiveness of these cells to microbial products, and to nonspecific stimulation (phorbol myristate acetate/ionomycin).4 The detected deficiency in CD4+ and CD8+ T-cell activation in this patient suggests that T cells reared in the absence of an intact APC compartment are anergic. Such an anergic T-cell phenotype was not observed in recently described patients with GATA2 haploinsufficiency who also develop deficiency of monocytes and DCs as well as progressive loss of B and NK lymphocytes, termed DC, monocyte, B and NK lymphoid deficiency.42,43 Although symptomatic GATA2-deficient patients also show impaired production of IL-12 and IFN-γ in whole blood assays, this may well reflect the absence of monocytes and NK cells rather than any T-cell autonomous defect. Furthermore, DC, monocyte, B and NK lymphoid-deficient patients can live well into adulthood before becoming ill,44 whereas the K108E patient was hospitalized before the age of 3 months4 and thus more strongly resembled infants with severe combined immunodeficiency who lack T-cell function altogether. This therefore highlights the uniqueness and severity of the congenital DC immunodeficiency associated with mutations in IRF8. Interestingly, we observed that the K108E patient had increased expression of genes and markers for mature B lymphocytes, and more specifically plasma cells (Figures 2C and 3C-D). This is in agreement with her hematologic profile showing increase in B cells compared with the normal values (Figure 1).3 In mice, Irf8 deficiency (Irf8−/−) has been shown to result in increased levels of mature marginal zone and follicular B cells in the spleen.45 Although this remains to be formally demonstrated, it is tempting to speculate that the role of IRF8 in restricting the expansion and maturation of B cells is conserved between mice and humans.
Using comparative RNA-seq and ChIP-seq, we further characterized the genes that are bound and regulated by IRF8 in human myeloid cells in vivo. This list of genes is presented in supplemental Table 2. These results support the interpretation that IRF8 functions primarily as a transcriptional activator in myeloid cells. This list is validated by the presence of well-known myeloid-specific genes and proteins (CD14, CD163, CD300e), as well as well-known proinflammatory molecules (IL-1β, CCL3, CCL4, C1QA) and their receptors (CCR5, CCR7, CSF1R, TGFBR2). The list also contains a number of other genes of unknown function that constitute novel IRF8 targets in human APCs. The study of the role of these poorly characterized genes in the ontogeny and function of myeloid cells promises to provide novel insight into the antimicrobial and immune function of these cells.
With respect to the pathological nature of the IRF8K108E mutation itself, we demonstrate a clear effect on IRF8 stability, which was unexpected. The decreased stability of the protein has pleiotropic effects on IRF8 targeting to the nucleus, posttranslational modification, and transactivation potential. These results strongly suggest that K108 plays an additional critical role in normal folding of the DBD of IRF8 in presence or in absence of a DNA template, and that its replacement to glutamic acid causes significant misfolding of the protein and increased proteasomal degradation following ubiquitination and SUMOylation of the misfolded variant. It is also possible that the K108R variant has a more subtle effect on IRF8 structure that may affect access to regulatory enzymes such as Cbl/DUBs (for ubiquitination) and E2I/SENP1 (for SUMOylation), thereby displacing the regulatory equilibrium between active IRF8 and inactive SUMOylated/ubiquitinated protein.33,41 Finally, studies in cells expressing both the K108E mutant and the WT protein demonstrate that K108E does not behave as a dominant negative and does not alter targeting of the WT protein to the nucleus. This is in agreement with the recessive mode of inheritance of the immunodeficiency and granulocytic hyperplasia phenotypes associated with the K108E mutation in this family.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Susan Gauthier for expert technical assistance.
This work was supported by a research grant from the National Institutes of Health, National Institute of Allergy and Infectious Diseases (R01AI035237) (P.G.), a James McGill Professorship salary award (P.G.), a fellowship from the Fonds de Recherche du Québec – Santé (D.L. and S.S.), a fellowship from the Canadian Institutes of Health Research Neuroinflammation training program (D.L.), and the Sir Jules Thorn Charitable Trust (S.H.).
Authorship
Contribution: S.S. performed all transactivation, nuclear localization, stability, ubiquitination, and SUMOylation assays; D.L. performed all RNA-seq–related experiments and bioinformatics analyses; F.L. performed bioinformatic analysis of RNA-seq data sets; G.B. supervised bioinformatic analysis of RNA-seq data sets; V.B. and M. H. obtained and handled initial clinical patient material; K.M.B. and T.R.L. conducted initial immunophenotyping of the patient material; A.B. and D.B. performed molecular modeling; J.-L.C. edited the manuscript; S.H. performed immunophenotyping of patient material and edited the manuscript; and P.G. designed and supervised the study and edited the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Philippe Gros, Department of Biochemistry and Complex Traits Group, McGill University, Bellini Life Sciences Building, Room 366, 3649 Sir William Osler Promenade, Montreal, QC H3G-0B1, Canada; e-mail philippe.gros@mcgill.ca.

![Figure 1. Blood cellular profile of patient K108E prior to and post stem cell transplant. Hematologic profiles were obtained by standard automated measurements (neutrophils, platelets, hemoglobin), and immune cell populations (total T lymphocytes, CD4+ T cells, CD8+ T cells, natural killer [NK] cells, B lymphocytes, monocytes) were established by fluorescence-activated cell sorter using cell-specific markers. (The degree of monocytopenia was inconsistently evident from machine differential counts beyond the patient’s initial presentation; those derived from flow cytometric evaluation are shown.) The patient was treated as hemophagocytic lymphohistiocytosis using a modified version of the HLH-2004 protocol (blue triangle) with resulting decline in neutrophil, B, and NK cell counts, accompanied by clinical stabilization. The patient then moved rapidly into conditioned hematopoietic stem cell transplant (red triangle; 9 months of age).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/124/12/10.1182_blood-2014-04-570879/4/m_1894f1.jpeg?Expires=1769092153&Signature=oIIeJOGIg1UcCLwJe3GCjstNJIDHHZPZQucvH4BCDF7oBVv0Hpn2WG5tbiLlw4BFNo8MUzWSPg9go2xTvCuUYFBoZpJ8F5t7Nu~ox0EobsIkjtJO5BOffVgZ2Wv-Wd8glmYxscqguRAnWYPgD08QcoYSYcxpnQxZA17q9Vg8pAe8tDKS0xWhNWN7ByjjEdbjz6vRas2oMSImnoZNZKhiAWi3TDwnVOhQWuJznlHwMPS7TuPRuaiLCNuTxsOArfOlOpZ0dfu83yKlfATUSjjdjAxM6FaPiatOwzwLnI~k8OrkG2OTadYwmIt3vwNDCdRNZessYeoANSchc~s~0NwdSA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal