Key Points
Myeloma cells are highly sensitive to PRIMA-1Met, independent of p53.
PRIMA-1Met induces myeloma cell death by impairing GSH/ROS balance.
Abstract
The aim of this study was to assess the efficiency of p53 reactivation and induction of massive apoptosis (PRIMA-1Met) in inducing myeloma cell death, using 27 human myeloma cell lines (HMCLs) and 23 primary samples. Measuring the lethal dose (LD50) of HMCLs revealed that HMCLs displayed heterogeneous sensitivity, with an LD50 ranging from 4 μM to more than 200 μM. The sensitivity of HMCLs did not correlate with myeloma genomic heterogeneity or TP53 status, and PRIMA-1Met did not induce or increase expression of the p53 target genes CDKN1A or TNFRSF10B/DR5. However, PRIMA-1Met increased expression of NOXA in a p53-independent manner, and NOXA silencing decreased PRIMA1Met-induced cell death. PRIMA-1Met depleted glutathione (GSH) content and induced reactive oxygen species production. The expression of GSH synthetase correlated with PRIMA-1Met LD50 values, and we showed that a GSH decrease mediated by GSH synthetase silencing or by and L-buthionine sulphoximine, an irreversible inhibitor of γ-glutamylcysteine synthetase, increased PRIMA-1Met-induced cell death and overcame PRIMA-1Met resistance. PRIMA-1Met (10 μM) induced cell death in 65% of primary cells independent of the presence of del17p; did not increase DR5 expression, arguing against an activation of p53 pathway; and synergized with L-buthionine sulphoximine in all samples. Finally, we showed in mouse TP53neg JJN3-xenograft model that PRIMA-1Met inhibited myeloma growth and synergized with L-buthionine sulphoximine in vivo.
Introduction
TP53 is the most frequently mutated gene in cancers, and the mutations are associated with resistance to therapy in numerous cancers, including multiple myeloma (MM).1,2 MM remains an incurable plasma cell malignancy, although treatments for the disease have progressed in the last decade.3 It is well established that at diagnosis, patients with a deletion of the short arm of chromosome 17 (del17p), which overlaps the TP53 locus (17p13), have been shown to have a shorter survival time, irrespective of the treatment regimens.3-7 Patients with del17p frequently (>30%) harbored a mutation in the remaining allele.8 Thus, there is an obvious need for compounds that bypass the defective p53 pathway, such as molecules targeting antiapoptotic molecules (eg, ABT-737 and ABT-199), which act downstream of p53.9,10 An alternative strategy is to target the overexpressed p53 mutant protein by directly modifying its conformation to restore its proapoptotic transcriptional functions.11 Indeed, molecules that can reactivate cell death in p53 mutant cells in a p53-dependent manner have been selected on the basis of their ability to either kill the cells (phenotypic screening) or bind to mutated p53 and restore a functional p53 conformation (biochemical screening).11-14 Among them, PRIMA-1Met (APR-246) was isolated according to its ability to restore apoptosis in SAOS2-His-273 cells in a p53-dependent manner and was shown to bind to p53.11,15,16 PRIMA-1Met, recently evaluated in a phase 1 study, was shown to be effective in vivo in 2 patients with a mutation in the core (V173M) or tetramerization domain (A355V). After treatment, expression of the p53 target genes Bax, Noxa, and Puma was increased in tumor cells.17
At a molecular level, p53 reactivation and induction of massive apoptosis (PRIMA-1Met) induced reactive oxygen species (ROS) production and requires an oxidative environment to reactivate p53, although the precise mechanism remains elusive.16 PRIMA-1Met may have other targets, such as oxidosqualene cyclase, which was identified using a docking-inverse approach.18 Several reports have described a p53-independent ability of PRIMA-1Met to induce tumor apoptosis. Indeed, in AML, APR-246 was found to be efficient irrespective of TP53 mutational status.19 Russo et al also have reported that PRIMA-1Met kills cell lines that lack p53 expression.20 Very recently, Saha et al reported that PRIMA-1Met kills MM cells via Noxa induction in a p53-independent manner.21
In the present work, we evaluated the efficacy of PRIMA-1Met across a collection of 27 human myeloma cell lines (HMCLs) representative of the heterogeneity of myeloma, as well as in 23 independent primary MM samples characterized for del17p. This HMCL collection was representative of TP53 abnormalities found in patients (chromosome 17p deletion, different point mutations, and exon deletion) and allowed us to provide an accurate preclinical evaluation.9,22-24 The efficacy of PRIMA-1Met was compared with that of nutlin3a, which reactivates the p53 pathway and induces cell death only in TP53wt HMCLs and primary cells.24 We show that PRIMA-1Met, in contrast to nutlin3a, killed HMCLs and primary myeloma cells independent of TP53 status and p53 expression in a GSH/ROS-dependent manner.
Materials and methods
HMCLs and primary myeloma cells
All HMCLs used in this article have been extensively characterized.9,23-26 The human stromal cell line HS5 was purchased from ATCC (LGC Standards SARL, Molsheim, France). After informed consent, blood or bone marrow samples from patients with MM were collected at the Department of Hematology at the University Hospital of Nantes or at the Intergroupe Francophone du Myélome (ethical approval DC-2011-1399). The study was conducted in accordance with the Declaration of Helsinki. Plasma cells were obtained after gradient density centrifugation, using Ficoll-Hypaque and purification with CD138 immunomagnetic beads (Stemcell Technologies, Le Plessis Robinson, France). Purified cells were cultured for 24 hours in RPMI1640 containing 5% fetal calf serum and 3 ng/mL interleukin 6. Deletion of the short arm of chromosome 17 was assessed by fluorescence in situ hybridization.8
Reagents and antibodies
Nutlin3a, N-acetyl-l-cysteine (NAC), glutathione reduced monoethyl ester (GSH-MEE), and l-buthionine sulphoximine (BSO) were purchased from Sigma-Aldrich (Saint-Quentin Fallavier, France). PRIMA-1Met, small interfering RNA (siRNA), and antibodies against caspase 2, caspase 3, γ-glutamate-cysteine synthetase modifier subunit (GCLM), and glutathione synthetase (GSS) were purchased from Santa Cruz Biotechnology (Clinisciences, Nanterre, France). Anti-Apo2.7-PE, anti-CD138-PE, control immunoglobulin G (IgG1)-phycoerythrin (PE) monoclonal antibodies (mAbs) were purchased from BD Biosciences (Le Pont de Claix, France), and anti-DR5-PE was purchased from eBioscience. Anti-p53, anti-Noxa, anti-Bax, anti-p21, and anti-actin antibodies were purchased from Oncogene Science (Life Technologies, Paris, France), Alexis Biotech (Enzo Life Sciences, Villeurbanne, France), Immunotech Beckman Coulter (Marseilles, France), Cell Signaling (Ozyme, St Quentin en Yvelines, France), and Millipore Bioscience Research Reagents (Molsheim, France), respectively. Glutathione (GSH) content was determined using a GSH/glutathione disulfide (GSSG) colorimetric detection kit (Arbor Assays, Euromedex, Strasbourg, France).
Cell death assays
ROS detection
Endogenous ROS production was assessed using CellROX oxidative stress reagent (Molecular Probes, Life Technologies, Saint-Aubin, France). CellROX Green reagent (5 μM) was added to the cell culture for 30 to 45 minutes at 37°C. After 3 washes in phosphate-buffered saline, fluorescence was analyzed on FACSCalibur (Cytocell, Structure Fédérative de Recherche Bonamy).
Gene silencing
Xenotransplant
Female severe combined immunodeficiency (SCID) beige 7-week-old mice were purchased from Charles River (L'Arbresle, France). Mice were bred and housed in the Experimental Therapeutic Unit (UTE, Structure Fédérative de Recherche Bonamy) under animal care license 44565. Tumors were generated by implanting 10 × 106 cells in 100 μL phosphate-buffered saline subcutaneously in the right flank above the hind leg.29 The tumor volume was recorded in 3 dimensions, using a digital caliper and calculated by measuring length × width × depth.
Statistical analysis
Statistical analyses were performed using the Kruskal-Wallis, analysis of variance, Fisher exact, Mann-Whitney, and Wilcoxon matched-pairs signed-rank tests.
Results
HMCLs displayed a p53-independent heterogeneous sensitivity to PRIMA-1Met
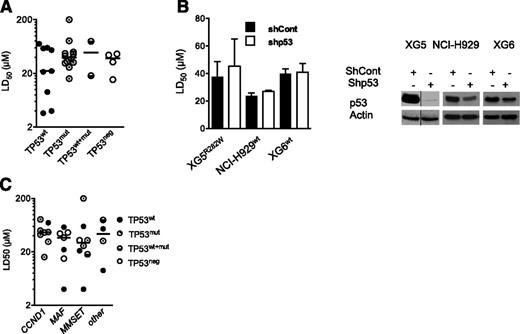
The present HMCL collection consisted of 9 TP53wt HMCLs, 12 TP53mut HMCLs, 2 TP53wt+mut, and 4 TP53neg HMCLs. TP53neg HMCLs did not express p53 protein because they either lacked p53 expression at the mRNA level (JJN3, KMS11) or had a premature stop codon (NAN1) or a lack of intron splicing (L363; supplemental Table 1, available on the Blood Web site).23,24 Thus, 4 HMCLs harbored a hot-spot mutated 175, 248, 273, or 282 codon. PRIMA-1Met LD50 values were determined by incubating myeloma cells for 72 hours with a serial dilution of PRIMA-1Met (1-200 μM) in culture media. A cell death assay was performed, using Apo2.7 staining.24,27 The LD50 values ranged from 3 up to more than 200 μM (median, 37 μM; supplemental Table 1). The median values for TP53wt, TP53mut, TP53wt+mut, and TP53neg HMCLs were, respectively, 22, 37, 49, and 38 μM, with no significant differences between the HMCLs (Figure 1A; P = .86). Myeloma cells lacking p53 expression, such as JJN3 and KMS11 (full length as well as all isoforms; data not shown), were as sensitive as TP53wt or TP53mut cells. Moreover, the shTP53 HMCLs, either TP53wt (XG6, NCI-H929) or TP53mut (XG5) myeloma cells, displayed no differential sensitivity to PRIMA-1Met (Figure 1B), confirming that PRIMA-1Met-induced myeloma cell death was not dependent on p53 expression. In contrast, the LD50 values for nutlin3a in shTP53 XG6 and NCI-H929 TP53wt HMCLs increased from 4 μM up to more than 10 μM (P < .05; supplemental Figure 1A), as described previously.24 Myeloma-specific genomic heterogeneity characterized by recurrent 14q32 translocation and the overexpression of the CCND1, MAF, or MMSET genes was not significantly associated with the PRIMA-1Met LD50 values (Figure 1C; P = .55).
Sensitivity of HMCLs to PRIMA-1Met was independent from TP53 status, p53 expression, and myeloma heterogeneity. (A) Sensitivity of HMCLs to PRIMA-1Met was independent of TP53 status. The LD50 values were defined by incubating cells (100 000 cells/0.2 mL) for 72 hours in the presence of a serial dilution of PRIMA-1Met (starting dose, 200 μM). Cell death was determined using Apo2.7 staining, as described previously.24,27 The LD50 values (defined as the mean of at least 3 independent experiments) were plotted against the TP53 status (supplemental Table 1). (B) Silencing of p53 in TP53wt (XG6, NCI-H929) and TP53R282W XG5 HMCLs did not inhibit PRIMA-1Met-induced cell death. Stable shTP53 HMCLs were previously reported.24 (Left) Cells were incubated for 72 hours, with serial dilutions of PRIMA-1Met (starting dose, 80 μM), and cell death was assessed as described in the legend of Figure 1A. The data represent the mean ± standard error of the mean (SEM) of 3 independent experiments. (Right) Western blot analysis of p53 expression in shCont and shp53 myeloma cells. (C) Sensitivity of HMCLs to PRIMA-1Met was independent of myeloma heterogeneity. The LD50 values were plotted against myeloma heterogeneity, characterized by recurrent 14q32, leading to overexpression of CCND1, (C-MAF or MAF-B) MAF, and MMSET.23
Sensitivity of HMCLs to PRIMA-1Met was independent from TP53 status, p53 expression, and myeloma heterogeneity. (A) Sensitivity of HMCLs to PRIMA-1Met was independent of TP53 status. The LD50 values were defined by incubating cells (100 000 cells/0.2 mL) for 72 hours in the presence of a serial dilution of PRIMA-1Met (starting dose, 200 μM). Cell death was determined using Apo2.7 staining, as described previously.24,27 The LD50 values (defined as the mean of at least 3 independent experiments) were plotted against the TP53 status (supplemental Table 1). (B) Silencing of p53 in TP53wt (XG6, NCI-H929) and TP53R282W XG5 HMCLs did not inhibit PRIMA-1Met-induced cell death. Stable shTP53 HMCLs were previously reported.24 (Left) Cells were incubated for 72 hours, with serial dilutions of PRIMA-1Met (starting dose, 80 μM), and cell death was assessed as described in the legend of Figure 1A. The data represent the mean ± standard error of the mean (SEM) of 3 independent experiments. (Right) Western blot analysis of p53 expression in shCont and shp53 myeloma cells. (C) Sensitivity of HMCLs to PRIMA-1Met was independent of myeloma heterogeneity. The LD50 values were plotted against myeloma heterogeneity, characterized by recurrent 14q32, leading to overexpression of CCND1, (C-MAF or MAF-B) MAF, and MMSET.23
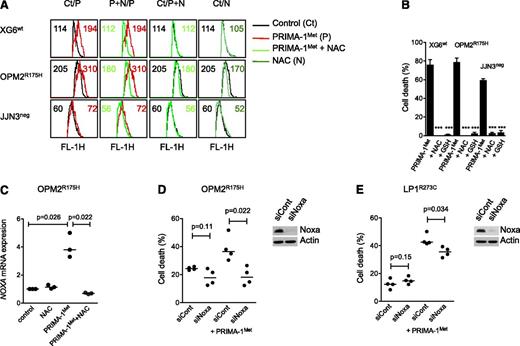
PRIMA-1Met did not increase expression of the p53 target genes p21 and DR5 but did increase the expression of Noxa
Despite a lack of correlation between sensitivity of myeloma cells to PRIMA-1Met and p53 expression, using flow cytometry or western blotting, we assessed whether PRIMA-1Met induced the expression of p53 target genes p21, Noxa, and DR5. We performed experiments using 3 HMCLs that expressed either a wild-type protein (XG6) or a TP53R175H mutant protein previously reported to be reactivated by PRIMA-1Met (OPM2) or no p53 protein (JJN3).16 Cells were treated with PRIMA-1Met (LD50 value) or nutlin3a (LD50 value or 10 μM) as a control of the p53 pathway mobilization. PRIMA-1Met did not induce p21 expression but did induce strong expression of Noxa in HMCLs, regardless of the p53 expression or status (Figure 2A). In contrast, nutlin3a increased p53, p21, and Noxa expression in XG6wt and NCI-H929wt cells (supplemental Figure 1B). Of note, the expression of p53, either mutated or wild-type, became undetectable after PRIMA-1Met treatment. PRIMA-1Met induced apoptosis, as revealed by the cleavage of caspases 2 and 3, and PARP. We previously reported that activation of a functional p53 pathway increased DR5 expression, as p53 directly bound to the DR5/TNFRSF10B gene.24 PRIMA-1Met did not increase DR5 expression; the median fold increases were 0.99 ± 0.09, 0.76 ± 0.25, and 0.89 ± 0.17 for XG6wt, OPM2R275H, and JJN3neg, respectively (Figure 2B). As expected, nutlin3a increased the expression of DR5 in XG6wt cells only (1.3 ± 0.1). These results confirmed that PRIMA-1Met-induced myeloma cell death occurred independent of p53.
PRIMA-1Met did not increase expression of the p53 target genes p21 and DR5 but did increase the expression of Noxa. (A) PRIMA-1Met did not induce the expression of the p53 target gene p21. XG6 (TP53wt), OPM2 (TP53R175H), and JJN3 (TP53neg) cells were treated overnight with 35 μM PRIMA-1Met, with or without 5 mM NAC. Protein expression was assessed using western blotting. (B) PRIMA-1Met did not increase the expression of the p53 target gene DR5. Cells, treated as described in the legend of Figure 2A, were strained with control-PE immunoglobulin or anti-DR5-PE mAbs, and fluorescence was analyzed on a FACsCalibur. The ratio of fluorescence was determined by dividing the specific fluorescence mean by the control fluorescence mean. Control staining of untreated and treated cells was identical and is not represented in the figure. The data represent 1 experiment out of 3.
PRIMA-1Met did not increase expression of the p53 target genes p21 and DR5 but did increase the expression of Noxa. (A) PRIMA-1Met did not induce the expression of the p53 target gene p21. XG6 (TP53wt), OPM2 (TP53R175H), and JJN3 (TP53neg) cells were treated overnight with 35 μM PRIMA-1Met, with or without 5 mM NAC. Protein expression was assessed using western blotting. (B) PRIMA-1Met did not increase the expression of the p53 target gene DR5. Cells, treated as described in the legend of Figure 2A, were strained with control-PE immunoglobulin or anti-DR5-PE mAbs, and fluorescence was analyzed on a FACsCalibur. The ratio of fluorescence was determined by dividing the specific fluorescence mean by the control fluorescence mean. Control staining of untreated and treated cells was identical and is not represented in the figure. The data represent 1 experiment out of 3.
Induction of Noxa and ROS was involved in myeloma cell death
Because PRIMA-1Met was reported to induce ROS, we assessed endogenous ROS production, using CellRox staining. XG6, OPM2, and JJN3 cells were incubated with 20 μM PRIMA-1Met, with or without the ROS scavenger NAC.16 Indeed, the mean fluorescence intensity was increased in cells treated with PRIMA-1Met, and this increase was inhibited by NAC (Figure 3A). The addition of NAC (5 mM) also inhibited Noxa increase, decrease in p53 expression, cleavage of both PARP and caspases, and cell death (Figures 2B and 3B). GSH, GSH-MEE (5 mM), also inhibited PRIMA-1Met-induced cell death in all sensitive HMCLs (P < .001; Figure 3B). In contrast, nutlin3a did not increase ROS production, and NAC did not impair nutlin3a-induced cell death in 3 TP53wt HMCLs (supplemental Figure 1C-D). The increase in Noxa expression was at the mRNA level: PRIMA-1Met increased the median expression of NOXA mRNA 3.8-fold in OPM2R175H cells (P = .026), and NAC fully prevented it (P = .022; Figure 3C). Silencing of Noxa inhibited PRIMA-1Met-induced cell death by 89% ± 21% (P = .022) in OPM2R175H and 36% ± 9% in LP1E286K cells (P = .034), respectively (Figure 3D-E).
Induction of ROS and Noxa was involved in myeloma cell death. (A) PRIMA-1Met induced ROS production. XG6 (TP53wt), OPM2 (TP53R175H), and JJN3 (TP53neg) cells (0.5 × 106/mL) were incubated overnight with 20 μM (JJN3) or 30 μM (XG6, OPM2) PRIMA-1Met in the presence or absence of 5 mM NAC. CellROX reagent (5 μM) was added to the cell culture for the last 30 minutes, at 37°C. Cells were washed in phosphate-buffered saline, and fluorescence was analyzed on FACSCalibur. (B) ROS scavengers NAC and GSH-MEE inhibited PRIMA-1Met-induced cell death. Cells were treated with PRIMA-1Met for 2 days in the presence or absence of NAC or GSH-MEE (5 mM). Cell death was assessed using Apo2.7 staining. The data represent the mean ± SEM of at least 3 experiments. ***P < .001. (C) PRIMA-1Met increased the expression of NOXA mRNA. OPM2 cells were treated overnight with 30 μM PRIMA-1Met in the presence or absence of NAC (5 mM). The expression of NOXA mRNA was performed using quantitative reverse-transcription polymerase chain reaction with the TaqMan probe. The data represent 3 independent experiments. (D-E) Transient silencing of Noxa inhibited PRIMA-1Met-induced cell death. OPM2 cells (B) or LP1 cells (C) were transfected with 100 pmol siCont or siNOXA (Life Technologies). At 72 hours, cells were treated for 24 hours with 100 μM PRIMA-1Met, and cell death was assessed using flow cytometry (Apo2.7 staining). The data represent 4 independent experiments. Noxa expression was assessed using western blotting 72 hours after siRNA transfection.
Induction of ROS and Noxa was involved in myeloma cell death. (A) PRIMA-1Met induced ROS production. XG6 (TP53wt), OPM2 (TP53R175H), and JJN3 (TP53neg) cells (0.5 × 106/mL) were incubated overnight with 20 μM (JJN3) or 30 μM (XG6, OPM2) PRIMA-1Met in the presence or absence of 5 mM NAC. CellROX reagent (5 μM) was added to the cell culture for the last 30 minutes, at 37°C. Cells were washed in phosphate-buffered saline, and fluorescence was analyzed on FACSCalibur. (B) ROS scavengers NAC and GSH-MEE inhibited PRIMA-1Met-induced cell death. Cells were treated with PRIMA-1Met for 2 days in the presence or absence of NAC or GSH-MEE (5 mM). Cell death was assessed using Apo2.7 staining. The data represent the mean ± SEM of at least 3 experiments. ***P < .001. (C) PRIMA-1Met increased the expression of NOXA mRNA. OPM2 cells were treated overnight with 30 μM PRIMA-1Met in the presence or absence of NAC (5 mM). The expression of NOXA mRNA was performed using quantitative reverse-transcription polymerase chain reaction with the TaqMan probe. The data represent 3 independent experiments. (D-E) Transient silencing of Noxa inhibited PRIMA-1Met-induced cell death. OPM2 cells (B) or LP1 cells (C) were transfected with 100 pmol siCont or siNOXA (Life Technologies). At 72 hours, cells were treated for 24 hours with 100 μM PRIMA-1Met, and cell death was assessed using flow cytometry (Apo2.7 staining). The data represent 4 independent experiments. Noxa expression was assessed using western blotting 72 hours after siRNA transfection.
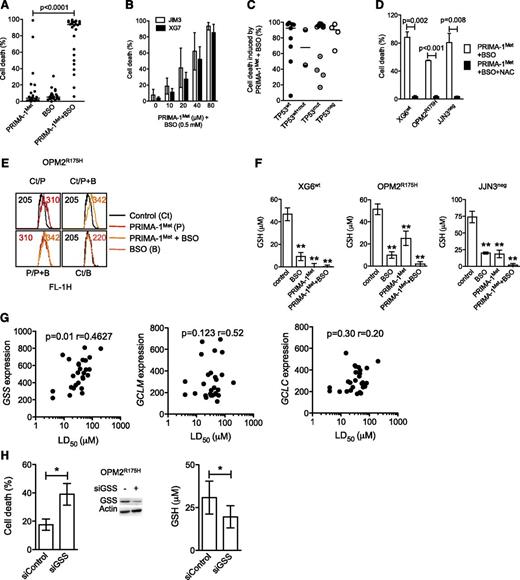
PRIMA-1Met impaired GSH metabolism
Because PRIMA-1Met induced ROS production, we assessed whether endogenous GSH depletion by BSO, an irreversible inhibitor of γ-glutamyl cysteine-synthase (γ-GCS), would favor PRIMA-1Met-induced cell death. Neither suboptimal concentration of PRIMA-1Met (10 μM) nor BSO (0.5 mM) alone induced any significant cell death (median, 4% and 6%, respectively) in 27 HMCLs, but they had significant synergy in all but XG7wt and JIM3R273S (median value, 93%; P < .0001; Figure 4A and supplemental Figure 2A). However, the synergy was recovered in XG7wt and JIM3R273S cells by increasing the doses of PRIMA-1Met (Figure 4B). The efficacy of the combination was not related to TP53 status (Figure 4C; P = .78), as confirmed in JJN3neg cells in which BSO increased PRIMA-1Met-induced caspase 3 activation and Noxa expression (supplemental Figure 2B). Addition of NAC also inhibited cell death induced by the combination of PRIMA-1Met with BSO by more than 90% in XG6, OPM2, and JJN3 (Figure 4D). Of note, the addition of BSO increased ROS production induced by PRIMA-1Met, as illustrated in OPM2 cells (Figure 4E). In contrast, BSO did not synergize with nutlin3a in TP53wt HMCLs (supplemental Figure 1E), nor did it overcome resistance to nutlin3a in OPM2R175H or JJN3neg cells (data not shown).
PRIMA-1Met impaired GSH metabolism in HMCLs. (A) BSO synergized with PRIMA-1Met. Cells were incubated for 48 hours with suboptimal doses of PRIMA-1Met (10 μM) and BSO (0.5 mM), and cell death was assessed using Apo2.7 staining. Each plot represents the mean cell death observed for each of the 27 HMCLs (obtained with 3 independent experiments). (B) Increasing doses of PRIMA-1Met restored synergy with BSO. XG7 and JIM3 cells were incubated for 48 hours with serial concentrations of PRIMA-1Met (10-80 μM) in the presence of 0.5 mM BSO. The data represent the mean ± SEM of 3 independent experiments. (C) Synergy between PRIMA-1Met and BSO was independent of TP53 status. Median cell death induced by PRIMA-1Met (10 μM) and BSO (0.5 mM) for each cell line was plotted against the TP53 status. (D) NAC overcame PRIMA-1Met and BSO synergy. Cells were incubated for 48 hours in the presence of PRIMA-1Met (10 μM) and BSO (0.5 mM), with or without NAC (5 mM). The data represent the mean ± SEM of 4 independent experiments. (E) BSO increased ROS production induced by PRIMA-1Met. OPM2 (0.5 × 106 cells/mL) was incubated overnight with 30 μM PRIMA-1Met or with 10 μM PRIMA-1Met and 0.5 mM BSO. CellROX reagent (5 μM) was added to the cell culture for the last 30 minutes, at 37°C. Cells were washed in phosphate-buffered saline, and fluorescence was analyzed on FACSCalibur. (F) PRIMA-1Met induced GSH depletion. Cells (0.5 × 106 cells/mL) were treated overnight with PRIMA-1Met alone (30 μM), BSO alone (0.5 mM), or a combination (10 μM and 0.5 mM). Cells were lysed in acidic buffer (10 × 106 cells/mL), and GSH content was determined using a colorimetric kit by following the instructions of the manufacturer. The data represent the mean ± SEM of 4 experiments. **P < .01. (G) Sensitivity of myeloma cells to PRIMA-1Met correlated with GSS expression. Expression of GSS or of the 2 subunits of γGCS (GCLM and GCLC) was plotted against LD50 PRIMA-1Met values. Correlation was assessed by Spearman test. (H) Silencing of GSS increased sensitivity to PRIMA-1Met. OPM2 cells were incubated for 3 days with siControl or siGSS RNA and treated with 25 μM PRIMA-1Met for the last 24 hours. Cell death was assessed by Apo2.7 staining. The data represent the mean ± SEM of 4 independent experiments. Expression of GSS (representative experiment) and GSH content (n = 4) was determined at 72 hours. *P < .05.
PRIMA-1Met impaired GSH metabolism in HMCLs. (A) BSO synergized with PRIMA-1Met. Cells were incubated for 48 hours with suboptimal doses of PRIMA-1Met (10 μM) and BSO (0.5 mM), and cell death was assessed using Apo2.7 staining. Each plot represents the mean cell death observed for each of the 27 HMCLs (obtained with 3 independent experiments). (B) Increasing doses of PRIMA-1Met restored synergy with BSO. XG7 and JIM3 cells were incubated for 48 hours with serial concentrations of PRIMA-1Met (10-80 μM) in the presence of 0.5 mM BSO. The data represent the mean ± SEM of 3 independent experiments. (C) Synergy between PRIMA-1Met and BSO was independent of TP53 status. Median cell death induced by PRIMA-1Met (10 μM) and BSO (0.5 mM) for each cell line was plotted against the TP53 status. (D) NAC overcame PRIMA-1Met and BSO synergy. Cells were incubated for 48 hours in the presence of PRIMA-1Met (10 μM) and BSO (0.5 mM), with or without NAC (5 mM). The data represent the mean ± SEM of 4 independent experiments. (E) BSO increased ROS production induced by PRIMA-1Met. OPM2 (0.5 × 106 cells/mL) was incubated overnight with 30 μM PRIMA-1Met or with 10 μM PRIMA-1Met and 0.5 mM BSO. CellROX reagent (5 μM) was added to the cell culture for the last 30 minutes, at 37°C. Cells were washed in phosphate-buffered saline, and fluorescence was analyzed on FACSCalibur. (F) PRIMA-1Met induced GSH depletion. Cells (0.5 × 106 cells/mL) were treated overnight with PRIMA-1Met alone (30 μM), BSO alone (0.5 mM), or a combination (10 μM and 0.5 mM). Cells were lysed in acidic buffer (10 × 106 cells/mL), and GSH content was determined using a colorimetric kit by following the instructions of the manufacturer. The data represent the mean ± SEM of 4 experiments. **P < .01. (G) Sensitivity of myeloma cells to PRIMA-1Met correlated with GSS expression. Expression of GSS or of the 2 subunits of γGCS (GCLM and GCLC) was plotted against LD50 PRIMA-1Met values. Correlation was assessed by Spearman test. (H) Silencing of GSS increased sensitivity to PRIMA-1Met. OPM2 cells were incubated for 3 days with siControl or siGSS RNA and treated with 25 μM PRIMA-1Met for the last 24 hours. Cell death was assessed by Apo2.7 staining. The data represent the mean ± SEM of 4 independent experiments. Expression of GSS (representative experiment) and GSH content (n = 4) was determined at 72 hours. *P < .05.
Because depletion of GSH induced by BSO synergized with PRIMA-1Met, we assessed whether PRIMA-1Met impaired GSH content. As shown in Figure 4F, PRIMA-1Met induced depletion in GSH content (GSH+GSSG). The decrease in GSH content induced by PRIMA-1Met was 52% ± 10%, 75% ± 8%, and 100% ± 5% in OPM2, JJN3, and XG6 cells, respectively. As expected, BSO decreased GSH content by 80% ± 4%, 81% ± 5%, and 73% ± 2% in XG6, OPM2, and JJN3, respectively (Figure 4F). Combination of PRIMA-1Met with BSO induced a total depletion in GSH content. In contrast, GSH content was slightly decreased (30% ± 10%) in nutlin3a-treated XG6 cells (supplemental Figure 1F).
Synthesis of GSH successively involves γ-GCS and GSS enzymes.30 PRIMA-1Met did not significantly modify expression of GCLM (regulatory subunit of γ-GCS) or GSS, indicating that GSH depletion was not caused by disappearance of any enzymes (supplemental Figure 3A). Expression of GSS, but not that of γGCS (GCLM or GCLC), was correlated with the LD50 PRIMA-1Met values (Figure 4G). Using siGSS RNA transfection, we showed that decrease in GSS expression in OPM2 cells increased their sensitivity to 25 μM PRIMA-1Met by twofold: cell death was 18% ± 4% and 39% ± 5% in siControl and siGSS cells, respectively (P = .026; Figure 4H), and GSH content was 31 ± 10 and 20 ± 6 μM in siControl and siGSS cells, respectively (P = .03).
HS5 stromal cells protected myeloma from PRIMA-1Met, but not from PRIMA-1Met plus BSO
Because the microenvironment can protect tumor cells, we assessed whether stromal cells could prevent PRIMA-1Met-induced myeloma cell death. Indeed, HS5 cells inhibited cell death induced by PRIMA-1Met by 50% in the 3 cell lines (P = .03), but they were unable to prevent PRIMA-1Met+BSO-induced cell death (supplemental Figure 3B). This inhibition was associated with a decrease in ROS production, in Noxa induction, and in GSH depletion (supplemental Figure 3C). Indeed, the mean global increase in GSH content induced by stromal cells was 1.23 ± 1.5, 2.0 ± 2.7 (P < .05), and 1.36 ± 0.5 in control, PRIMA-1Met, and PRIMA-1Met+BSO-treated cell lines (supplemental Figure 3D). In contrast, stromal cells did not protect XG6 from nutlin3a-induced cell death (supplemental Figure 3G).
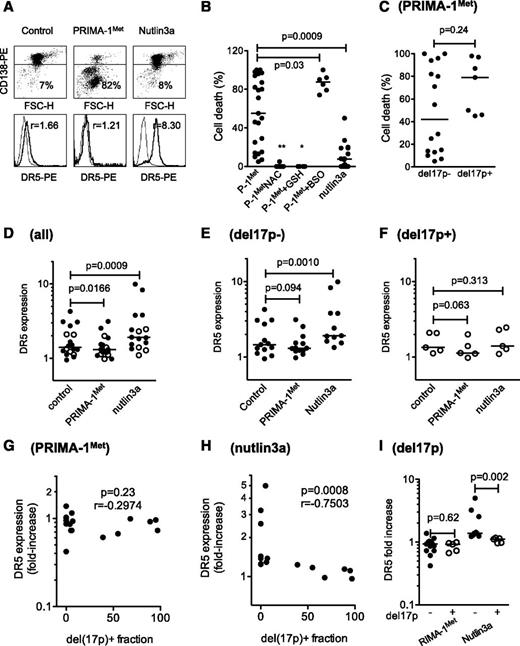
PRIMA-1Met induced cell death in primary cells irrespective of del17p status and synergized with BSO
The effect of PRIMA-1Met was further assessed using primary cells (25 samples from 23 consecutive patients). Cell death was assessed by the loss of CD138 expression, and modulation of DR5 expression was used as a surrogate for the activation of the p53 pathway, as previously described and illustrated in Figure 5A.24 PRIMA-1Met, which significantly reduced myeloma survival (P = .0067 Wilcoxon matched-pairs signed rank test), induced a median cell death of 55% (Figure 5B and Table 1). The sensitivity of myeloma cells taken from PCL or MM patients was not significantly different (P = .34). Although NAC (n = 9) or GSH-MEE (n = 6) inhibited cell death (P = .009 and P = .036, respectively), the addition of BSO increased PRIMA-1Met-induced cell death (87.5% of median cell death vs 32% with PRIMA-1Met alone; P = .037; n = 6; and 0% with BSO alone). PRIMA-1Met was more efficient than nutlin3a at inducing myeloma cell death (55% vs 7%, respectively; Figure 5B). The presence of a hemi-deletion of the 17p chromosome (del17p) negatively impairs overall patient survival, regardless of treatment, and TP53 mutations were found in samples harboring a del17p.8 Samples displayed a 17p deletion in a minor (<50%, considered as del17p−) or major (>50%, considered as del17p+) fraction of cells. Cell death induced by PRIMA-1Met was not different in samples with or without del17p (P = .24, Mann-Whitney test; Figure 5C). PRIMA-1Met induced a decrease in DR5 expression in primary samples (median fold increase, 0.92; n = 18; P = .0166; Figure 5D), whether harboring a del17p or not (median fold increase, 0.93 [n = 5; P = .0625], and median fold increase, 0.93 [n = 11; P = .0942], respectively; Figure 5E-F). Moreover, no correlation was found between the proportion of cells with del17p and the modulation in DR5 expression (P = .23, Spearman test; Figure 5G). In contrast, nutlin3a induced a significant increase in DR5 expression (median value, 1.25; n = 16; P < .001), which was significant in samples without del17p (median fold increase, 1.38; n = 11; P = .001), but not in samples with del17p (median fold increase, 1.10; n = 5; P = .31; Figure 5D-F). The increase in DR5 expression induced by nutlin3a was negatively correlated with the proportion of del17p within the samples (P < .001; Figure 5H). Finally, the increase in DR5 expression induced by nutlin-3a, but not by PRIMA-1Met, was significantly higher in samples without del17p (Figure 5I). Thus, in primary myeloma cells, PRIMA-1Met, in contrast to nutlin3a, did not increase DR5 expression and, instead, induced cell death irrespective of del17p status in synergy with BSO.
PRIMA-1Met induced cell death in primary cells, irrespective of TP53 status, and synergized with BSO. (A) PRIMA-1Met induced cell death in primary cells. Purified myeloma cells (sample 10; Table 1) were treated overnight with PRIMA-1Met (10 μM) or nutlin3a (10 μM) and stained with CD138-PE (cytogram), control-PE, or anti-DR5-PE (histogram) mAbs to assess cell death (loss of CD138 staining) or DR5 expression level. The thin line represents control staining, and the thick line is DR5 staining. (B) PRIMA-1Met induced primary myeloma cell death and synergized with BSO but was antagonized by NAC or GSH-MEE. Primary myeloma cells were isolated from the bone marrow or peripheral blood of patients with MM or plasma cell leukemia. Cells were incubated overnight in the presence or absence of PRIMA-1Met (10 μM), BSO (0.5 mM), NAC (5 mM), GSH-MEE (5 mM), or nutlin3a (10 μM). BSO, NAC, and GSH-MEE did not induce any myeloma cell death (data not shown). Cell death was assessed by the loss of CD138 staining, as described previously. (C) PRIMA-1Met-induced cell death was independent of del17p status. The TP53 status of primary cells was assessed, using fluorescence in situ hybridization, by defining the percentage of myeloma cells lacking the short arm of chromosome 17 (del17p), as previously reported.24 In this cohort, deletion was considered negative when a minority of cells harbored a 17p deletion (range, 0%-39%), and positive when more than 50% of cells harbored the deletion (range, 50%-97%; Table 1). Samples 6′ and 22′ were excluded from this analysis to analyze independent samples. (D-F) PRIMA-1Met, in contrast to nutlin3a, did not increase DR5 expression in primary cells. Cells were treated overnight with PRIMA-1Met (10 μM) or nutlin3a (10 μM) and were stained with control-PE or anti-DR5-PE mAbs. DR5 expression was calculated by dividing the specific mean fluorescence (DR5) of untreated and treated cells by the control staining of untreated and treated cells, respectively. The variation of DR5 expression was analyzed in the whole samples (D) and in samples without (E) or with (F) del17p. (G) Modulation in DR5 expression induced by PRIMA-1Met was not correlated with del17. The fold-increase in DR5 expression was plotted against the percentage of cells with del17p within the samples. (H) Modulation in DR5 expression induced by nutlin3a was inversely correlated to del17p. The fold-increase in DR5 expression was plotted against the percentage of cells with del17p within the samples. (I) Modulation in DR5 expression induced by nutlin3a, but not by PRIMA-1Met, was significantly higher in samples without del17p than in samples with del17p. *P < .05; ***P < .001.
PRIMA-1Met induced cell death in primary cells, irrespective of TP53 status, and synergized with BSO. (A) PRIMA-1Met induced cell death in primary cells. Purified myeloma cells (sample 10; Table 1) were treated overnight with PRIMA-1Met (10 μM) or nutlin3a (10 μM) and stained with CD138-PE (cytogram), control-PE, or anti-DR5-PE (histogram) mAbs to assess cell death (loss of CD138 staining) or DR5 expression level. The thin line represents control staining, and the thick line is DR5 staining. (B) PRIMA-1Met induced primary myeloma cell death and synergized with BSO but was antagonized by NAC or GSH-MEE. Primary myeloma cells were isolated from the bone marrow or peripheral blood of patients with MM or plasma cell leukemia. Cells were incubated overnight in the presence or absence of PRIMA-1Met (10 μM), BSO (0.5 mM), NAC (5 mM), GSH-MEE (5 mM), or nutlin3a (10 μM). BSO, NAC, and GSH-MEE did not induce any myeloma cell death (data not shown). Cell death was assessed by the loss of CD138 staining, as described previously. (C) PRIMA-1Met-induced cell death was independent of del17p status. The TP53 status of primary cells was assessed, using fluorescence in situ hybridization, by defining the percentage of myeloma cells lacking the short arm of chromosome 17 (del17p), as previously reported.24 In this cohort, deletion was considered negative when a minority of cells harbored a 17p deletion (range, 0%-39%), and positive when more than 50% of cells harbored the deletion (range, 50%-97%; Table 1). Samples 6′ and 22′ were excluded from this analysis to analyze independent samples. (D-F) PRIMA-1Met, in contrast to nutlin3a, did not increase DR5 expression in primary cells. Cells were treated overnight with PRIMA-1Met (10 μM) or nutlin3a (10 μM) and were stained with control-PE or anti-DR5-PE mAbs. DR5 expression was calculated by dividing the specific mean fluorescence (DR5) of untreated and treated cells by the control staining of untreated and treated cells, respectively. The variation of DR5 expression was analyzed in the whole samples (D) and in samples without (E) or with (F) del17p. (G) Modulation in DR5 expression induced by PRIMA-1Met was not correlated with del17. The fold-increase in DR5 expression was plotted against the percentage of cells with del17p within the samples. (H) Modulation in DR5 expression induced by nutlin3a was inversely correlated to del17p. The fold-increase in DR5 expression was plotted against the percentage of cells with del17p within the samples. (I) Modulation in DR5 expression induced by nutlin3a, but not by PRIMA-1Met, was significantly higher in samples without del17p than in samples with del17p. *P < .05; ***P < .001.
PRIMA-1Met induced cell death in primary cells
| Sample . | Patient’s characteristics . | Cell death . | Ratio of DR5 expression . | DR5 fold increase . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease . | Status . | Origin . | del17p . | PRIMA1Me . | PRIMA1Met+NAC . | PRIMA1Met+GSH . | PRIMA1Met+BSO . | Nutlin3a . | Control . | PRIMA1Met . | Nutlin3a . | PRIMA1Met . | Nutlin3a . | |
| 1 | pPCL | D | PB | 0% | 80% | — | — | — | 0% | 0.95 | 1.31 | 2.42 | 1.38 | 2.55 |
| 2 | MM | R | BM | 0% | 71% | — | — | — | 0% | 1.46 | 1.51 | 1.83 | 1.03 | 1.25 |
| 3 | sPCL | R | PB | 0% | 29% | 5% | — | — | 19% | 1.31 | 1.32 | 1.62 | 1.01 | 1.24 |
| 4 | sPCL | R | PB | 0% | 13% | 0% | 0% | 92% | 2% | 1.51 | 1.54 | 1.89 | 1.02 | 1.25 |
| 5 | MM | R | BM | 0% | 100% | 0% | 0% | 100% | 0% | 3.07 | 1.30 | 9.92 | 0.42 | 3.23 |
| 6 | pPCL | D | BM | 0% | 14% | 0% | 0% | 74% | 0% | — | — | — | — | — |
| 6’ | pPCL | D | PB | 0% | 6% | 0% | 0% | 98% | 0% | — | — | — | — | — |
| 7 | sPCL | R | PB | 0% | 5% | 0% | 0% | 85% | 50% | — | — | — | — | — |
| 8 | MM | D | BM | 0% | 79% | — | — | — | 0% | 1.12 | 0.98 | 1.54 | 0.87 | 1.38 |
| 9 | MM | D | BM | 0% | 46% | — | — | — | 0% | 1.34 | 1.23 | 1.92 | 0.95 | 1.43 |
| 10 | MM | D | BM | 4% | 100% | 0% | 0% | — | — | 1.26 | 1.09 | — | 0.86 | |
| 11 | MM | D | BM | 4% | 15% | 0% | — | — | — | — | — | — | — | — |
| 12 | MM | D | BM | 5% | 82% | — | — | — | 8% | 1.66 | 1.21 | 8.30 | 0.73 | 5.00 |
| 13 | pPCL | D | BM | 6% | 25% | — | — | — | 20% | 1.04 | 1.20 | 1.34 | 1.15 | 1.29 |
| 14 | MM | D | BM | 6% | 10% | 0% | 0% | — | — | 4.27 | 3.13 | — | 0.73 | |
| 15 | MM | D | BM | 7% | 7% | — | — | — | 13% | 2.73 | 2.54 | 3.78 | 0.93 | 1.38 |
| 16 | MM | R | BM | 39% | 94% | — | — | — | 8% | 3.10 | 1.88 | 3.81 | 0.61 | 1.23 |
| 17 | MM | D | BM | 55% | 98% | — | — | — | — | 2.10 | 1.40 | 2.46 | 0.67 | 1.17 |
| 18 | sPCL | R | PB | 68% | 45% | — | — | — | 27% | 1.12 | 1.11 | 1.10 | 0.99 | 0.98 |
| 19 | pPCL | D | PB | 89% | 87% | — | — | 90% | 19% | 1.23 | 1.13 | 1.40 | 0.92 | 1.14 |
| 20 | MM | D | BM | 90% | 55% | — | — | — | 1% | — | — | — | — | — |
| 21 | sPCL | R | PB | 95% | 97% | — | — | — | 7% | 2.08 | 2.00 | 2.30 | 0.96 | 1.11 |
| 22 | pPCL | D | BM | 96% | 50% | 0% | — | 79% | — | — | — | — | — | — |
| 22’ | pPCL | D | PB | 96% | 51% | 0% | — | 96% | — | — | — | — | — | — |
| 23 | pPCL | D | PB | 97% | 96% | — | — | — | 11% | 1.34 | 0.98 | 1.28 | 0.73 | 0.96 |
| Sample . | Patient’s characteristics . | Cell death . | Ratio of DR5 expression . | DR5 fold increase . | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease . | Status . | Origin . | del17p . | PRIMA1Me . | PRIMA1Met+NAC . | PRIMA1Met+GSH . | PRIMA1Met+BSO . | Nutlin3a . | Control . | PRIMA1Met . | Nutlin3a . | PRIMA1Met . | Nutlin3a . | |
| 1 | pPCL | D | PB | 0% | 80% | — | — | — | 0% | 0.95 | 1.31 | 2.42 | 1.38 | 2.55 |
| 2 | MM | R | BM | 0% | 71% | — | — | — | 0% | 1.46 | 1.51 | 1.83 | 1.03 | 1.25 |
| 3 | sPCL | R | PB | 0% | 29% | 5% | — | — | 19% | 1.31 | 1.32 | 1.62 | 1.01 | 1.24 |
| 4 | sPCL | R | PB | 0% | 13% | 0% | 0% | 92% | 2% | 1.51 | 1.54 | 1.89 | 1.02 | 1.25 |
| 5 | MM | R | BM | 0% | 100% | 0% | 0% | 100% | 0% | 3.07 | 1.30 | 9.92 | 0.42 | 3.23 |
| 6 | pPCL | D | BM | 0% | 14% | 0% | 0% | 74% | 0% | — | — | — | — | — |
| 6’ | pPCL | D | PB | 0% | 6% | 0% | 0% | 98% | 0% | — | — | — | — | — |
| 7 | sPCL | R | PB | 0% | 5% | 0% | 0% | 85% | 50% | — | — | — | — | — |
| 8 | MM | D | BM | 0% | 79% | — | — | — | 0% | 1.12 | 0.98 | 1.54 | 0.87 | 1.38 |
| 9 | MM | D | BM | 0% | 46% | — | — | — | 0% | 1.34 | 1.23 | 1.92 | 0.95 | 1.43 |
| 10 | MM | D | BM | 4% | 100% | 0% | 0% | — | — | 1.26 | 1.09 | — | 0.86 | |
| 11 | MM | D | BM | 4% | 15% | 0% | — | — | — | — | — | — | — | — |
| 12 | MM | D | BM | 5% | 82% | — | — | — | 8% | 1.66 | 1.21 | 8.30 | 0.73 | 5.00 |
| 13 | pPCL | D | BM | 6% | 25% | — | — | — | 20% | 1.04 | 1.20 | 1.34 | 1.15 | 1.29 |
| 14 | MM | D | BM | 6% | 10% | 0% | 0% | — | — | 4.27 | 3.13 | — | 0.73 | |
| 15 | MM | D | BM | 7% | 7% | — | — | — | 13% | 2.73 | 2.54 | 3.78 | 0.93 | 1.38 |
| 16 | MM | R | BM | 39% | 94% | — | — | — | 8% | 3.10 | 1.88 | 3.81 | 0.61 | 1.23 |
| 17 | MM | D | BM | 55% | 98% | — | — | — | — | 2.10 | 1.40 | 2.46 | 0.67 | 1.17 |
| 18 | sPCL | R | PB | 68% | 45% | — | — | — | 27% | 1.12 | 1.11 | 1.10 | 0.99 | 0.98 |
| 19 | pPCL | D | PB | 89% | 87% | — | — | 90% | 19% | 1.23 | 1.13 | 1.40 | 0.92 | 1.14 |
| 20 | MM | D | BM | 90% | 55% | — | — | — | 1% | — | — | — | — | — |
| 21 | sPCL | R | PB | 95% | 97% | — | — | — | 7% | 2.08 | 2.00 | 2.30 | 0.96 | 1.11 |
| 22 | pPCL | D | BM | 96% | 50% | 0% | — | 79% | — | — | — | — | — | — |
| 22’ | pPCL | D | PB | 96% | 51% | 0% | — | 96% | — | — | — | — | — | — |
| 23 | pPCL | D | PB | 97% | 96% | — | — | — | 11% | 1.34 | 0.98 | 1.28 | 0.73 | 0.96 |
Primary myeloma cells were purified (samples 2, 5, 6, 8, 9, 11, 12, 14, 15, 16, 17, 20, 22, and 23) or not (samples 1, 3, 4, 7, 10, 13, 18, 19, and 21), with CD138-conjugated magnetic beads, and incubated overnight in RPMI1640 supplemented with 5% fetal calf serum and 3 ng/mL interleukin 6, with or without nutlin3a (10 μM), PRIMA-1Met (10 μM), NAC (5 mM), GSH (5 mM), or BSO (0.5 mM), as indicated. Cell death was assessed by the loss of CD138 staining. Levels of DR5 expression were assessed by direct staining with anti-DR5-PE or control-PE mAb of untreated (control) or PRIMA-1Met- or nutlin3a-treated cells, and was expressed as the ratio of the specific fluorescence mean to that of control staining in untreated and treated cells. Samples 6′ and 22′ were peripheral blood samples from patients 6 and 22 and were not considered for statistical analyses. Dash indicates not done. BM, bone marrow; D, diagnosis; p, primary; PB, peripheral blood; PCL, plasma cell leukemia; R, relapse; s, secondary.
PRIMA-1Met synergized with BSO in vivo in a xenograft model
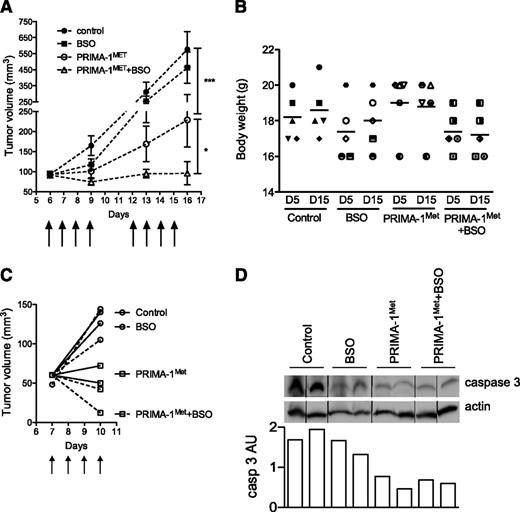
We assessed PRIMA-1Met efficiency at preventing myeloma growth, both alone and in combination with BSO. SCID-beige mice bearing JJN3 tumor cells received either no treatment (control), PRIMA-1Met (18 mg/kg, intravenous injection), BSO (10 mM, drinking water), or a combination of BSO and PRIMA-1Met. PRIMA-1Met (20 mg/kg) was previously shown to be efficient in preventing SAOS2 growth in vivo.15 BSO (30 mM) was evaluated in combination with melphalan.31 Mice were treated with 10 mM BSO in drinking water because this concentration was shown to reduce GSH content in the liver.32 Treatments were performed daily for 4 days, stopped for 2 days, and then performed again for another 4 days (as indicated by arrows in Figure 6A). Mice were then killed at day 16 because control and BSO-treated tumors exceeded the authorized tumor load. Body weight was not significantly affected by any treatments: The variation of weight between day 5 and day 15 was 102.2% ± 3.0%, 102.4% ± 3.3%, 99.0% ± 2.2%, and 98.8% ± 2.7% for control, BSO-treated, PRIMA-1Met-treated, and PRIMA-1Met+BSO-treated mice, respectively (Figure 6B). The presence of BSO did not significantly modify tumor growth (P > .05, 2-way analysis of variance test). In contrast, PRIMA-1Met significantly impaired tumor growth (P < .001), and its combination with BSO further inhibited tumor growth (P < .05). Tumor volume increased by 6.2-, 5.0-, 2.5-, and 1.05-fold in control, BSO-treated, PRIMA-1Met-treated, and PRIMA-1Met+BSO-treated mice, respectively. Tumor size tended to increase during treatment with PRIMA-1Met; the mean tumor size was 93.0 ± 7.4 mm3 and 228.8 ± 67.3 mm3 at day 6 and day 16, respectively (P = .18, Wilcoxon matched-pairs signed rank test). In contrast, in the presence of PRIMA-1Met and BSO, the tumors failed to increase in size; mean tumor volume was 91.4 ± 5.9 mm3 and 106 ± 30.6 mm3 at day 6 and day 16, respectively (P = .7). In another set of mice, tumors were removed after 4 days of treatment to analyze expression of caspase 3 (Figure 6C). The level of procaspase 3 was decreased in tumors from mice treated with PRIMA-1Met±BSO (Figure 6D).
PRIMA-1Met synergized with BSO in vivo. (A) PRIMA-1Met and PRIMA-1Met+BSO treatment inhibited in vivo tumor growth. Seven-week-old female SCID-beige mice were subcutaneously injected with 10 × 106 JJN3 cells. After 6 days, 20 mice bearing similar tumor loads were selected and randomly divided into 4 groups receiving no treatment (control), 18 mg/kg PRIMA-1MET (intravenous injection), 10 mM BSO (drinking water), or 18 mg/kg PRIMA-1MET and 10 mM BSO. Treatment was performed daily for 4 days, stopped, and then resumed for 4 additional days. Mice were killed the day after the final treatment (day 15). The tumor volume was assessed at days 6, 9, 13, and 16. *P < .05; ***P < .001. (B) PRIMA-1Met and PRIMA-1Met+BSO did not significantly alter body weight. The figure represents the body weights assessed at days 5 and 15 for each mouse (1 symbol per mouse). (C-D) PRIMA-1Met and PRIMA-1Met + BSO treatment increased caspase 3 activity. Eight mice were subcutaneously injected with 10 × 106 JJN3 cells. After 7 days, mice received no treatment (control), 18 mg/kg PRIMA-1MET (intravenous injection), 10 mM BSO (drinking water), or 18 mg/kg PRIMA-1MET and 10 mM BSO over the course of 4 days. Mice were killed 4 hours after the last treatment, and tumors were disrupted with a dounce homogenizer in 0.4% Triton X-100. Expression of proteins was analyzed by western blotting. Expression of procaspase 3 (arbitrary unit) was normalized, using actin expression.
PRIMA-1Met synergized with BSO in vivo. (A) PRIMA-1Met and PRIMA-1Met+BSO treatment inhibited in vivo tumor growth. Seven-week-old female SCID-beige mice were subcutaneously injected with 10 × 106 JJN3 cells. After 6 days, 20 mice bearing similar tumor loads were selected and randomly divided into 4 groups receiving no treatment (control), 18 mg/kg PRIMA-1MET (intravenous injection), 10 mM BSO (drinking water), or 18 mg/kg PRIMA-1MET and 10 mM BSO. Treatment was performed daily for 4 days, stopped, and then resumed for 4 additional days. Mice were killed the day after the final treatment (day 15). The tumor volume was assessed at days 6, 9, 13, and 16. *P < .05; ***P < .001. (B) PRIMA-1Met and PRIMA-1Met+BSO did not significantly alter body weight. The figure represents the body weights assessed at days 5 and 15 for each mouse (1 symbol per mouse). (C-D) PRIMA-1Met and PRIMA-1Met + BSO treatment increased caspase 3 activity. Eight mice were subcutaneously injected with 10 × 106 JJN3 cells. After 7 days, mice received no treatment (control), 18 mg/kg PRIMA-1MET (intravenous injection), 10 mM BSO (drinking water), or 18 mg/kg PRIMA-1MET and 10 mM BSO over the course of 4 days. Mice were killed 4 hours after the last treatment, and tumors were disrupted with a dounce homogenizer in 0.4% Triton X-100. Expression of proteins was analyzed by western blotting. Expression of procaspase 3 (arbitrary unit) was normalized, using actin expression.
Discussion
In this study, we assessed the efficacy of PRIMA-1Met, a p53 targeted drug selected for its ability to reactivate mutant p53 proteins, in inducing apoptosis in myeloma cell lines (n = 27) and primary myeloma cells (n = 23), both in vitro and in vivo. The TP53 status of HMCLs and primary cells was assessed by direct sequencing of reverse transcription-polymerase chain reaction products and by the presence of deletion of the short arm of chromosome 17, del17p, as previously described.24 We showed that PRIMA-1Met induced myeloma cell death, irrespective of TP53 status. Moreover, HMCLs lacking p53 expression (as well as p53 isoforms, data not shown), such as JJN3 or KMS11, were as sensitive as TP53mut or TP53wt HMCLs. In primary cells, we showed that 10 μM PRIMA-1Met induced cell death in primary samples (median cell death, 50%) without significant differences between samples with or without del17p, and PRIMA-1Met, in contrast to nutlin3a, did not induce any increase in DR5 expression. Using 3 HMCLs expressing either a wild-type p53 protein (XG6) or a R175H mutant protein harboring a mutation that could be reactivated by PRIMA-1Met (OPM2), or expressing no p53 protein (JJN3), we showed that PRIMA-1Met did not reactivate p53 target genes such as CDKN1A or TNFRSF10B but, rather, increased expression of NOXA and induced production of ROS. In the presence of the ROS scavengers NAC or GSH-MEE, cell death, Noxa expression, and ROS production induced by PRIMA-1Met were prevented. In contrast, NAC did not prevent cell death or Noxa increase induced by nutlin3a. We show that PRIMA-1Met, but not nutlin3a, induced the depletion of GSH content. Moreover, irreversible inhibition of γGCS by BSO synergized with PRIMA-1Met in cell lines, in primary samples, and in vivo. Involvement of GSH metabolism was further confirmed in GSS-silenced cells, which became more sensitive to PRIMA-1Met. BSO did not synergize with nutlin3a or RITA (another MDM2/p53 inhibitor, data not shown), which confirms that the mechanism of cell death induced by PRIMA-1Met is different from that induced by p53 stabilization.
Similar data showing the ability of PRIMA-1Met to kill tumor cells independent of p53 were recently reported in several tumor models, including myeloma.19-21 Of note, PRIMA-1Met was isolated from the chemical library by a phenotypic screening that assessed the alternative sensitivity of Saos-2-His-273 cells carrying tetracycline-regulated mutant p53.33 In contrast, nutlin3a was isolated in a biochemical screen via its direct binding to MDM2.34 These different screening strategies may, at least in part, explain why PRIMA-1Met has activities beyond binding to p53. Although PRIMA-1Met was not specifically linked to p53, the drug was able to kill myeloma cells such as OPM2 or JJN3, which are resistant to the alkylating drugs melphalan and bendamustin.26 Of major interest, Peng et al recently reported that PRIMA-1Met targeted the selenoprotein thioredoxin reductase 1 and converted it into a NADPH oxidase enzyme.35 Our data show that PRIMA-1Met induced a depletion of GSH content (GSH+GSSG), suggesting an inhibition of GSH synthesis. Moreover, silencing of GSS increased sensitivity of OPM2 cells to PRIMA-1Met, confirming that GSH plays a central role.
Because NAC or GSH-MEE can, either indirectly via γGCS or directly, increase intracellular GSH content, their ability to block PRIMA-1Met-induced cell death could be related to an increase in GSH content. Indeed, PRIMA-1Met-induced cell death was fully prevented by NAC or GSH-MEE, as well as partly by another ROS scavenger, L-ascorbic acid (50% of inhibition, data not shown). However, because NAC overcame the BSO-PRIMA-1Met synergy and because BSO is an irreversible inhibitor or γGCS, NAC cannot only act as a source of GSH. This suggests that beyond their ability to neutralize ROS and regenerate GSH, NAC or GSH-MEE can directly interact with PRIMA-1Met to neutralize it, as shown by Lambert et al.16 The depletion of GSH by BSO did not induce cell death and did not induce ROS production. Thus, PRIMA-1Met-induced GSH depletion is necessary but not sufficient to induce cell death. Indeed, because PRIMA-1Met can convert the antioxidant activity of thioredoxin into a prooxidant activity and because thioredoxin reductase and GSH peroxidase are essential to neutralize ROS, it appears that PRIMA-1Met-treated cells do not have any functional detoxifying metabolism and cannot face ROS.
The mechanism of increase in Noxa expression induced by PRIMA-1Met remains unclear. Recently, Saha et al reported that PRIMA-1Met-induced cell death was mediated by Noxa, via p73 in myeloma cell lines.21 Although we also showed that Noxa silencing in OPM2 or LP1 cells decreased cell death induced by PRIMA-1Met, we failed to find any increase in p73 expression (data not shown) or in p53/p73 target genes such as p21. P73 is not constitutively expressed in myeloma cells because of its silencing by DNA methylation, and it seems unlikely that PRIMA-1Met inhibits DNA methylation within 24 hours in cell lines or in primary cells, which do not proliferate.36 Myeloma cells are sensitive to ROS, and ROS production is involved in melphalan or bendamustine-induced myeloma cell death. Indeed, although NAC and GSH partly prevent melphalan-induced tumor death, BSO synergizes with melphalan.26,37-39 The melphalan plus BSO combination was also assessed in phase 1 studies, which reported that the toxicity was acceptable.40,41 However, no phase 3 study is available to determine the ultimate efficacy of that combination. We show in the JJN3-SCID xenograft model that 18 mg/kg PRIMA-1Met was enough to inhibit tumor growth and that addition of BSO induced tumor regression. PRIMA-1Met (100 mg/kg) was also shown to reduce tumor growth in the RPMI8226 xenograft model and to synergize with dexamethasone or doxorubicine.21 In both RPMI8226 and JJN3 xenograft models, mouse body weight remained stable, excluding the possibility of major toxicity from PRIMA-1Met. Thus, evaluating the combined results, it appears that PRIMA-1Met is effective against myeloma cells, both in vitro and in vivo, and that its efficacy is increased when it is used in combination with myeloma-approved drugs or BSO.
In JJN3-SCID mice, administration of PRIMA-1Met was performed to mimic the reported assessment of PRIMA-1Met/APR-246 in patients who received APR-246 once per day for 4 consecutive days over the course of 1 or 2 weeks.17 This phase 1 study, which involved patients with prostate cancer and hematologic malignancies, including myeloma, established that the maximum tolerated dose is 60 mg/kg. The authors reported that the tumor load of 2 patients with TP53mut tumors decreased during treatment. Our in vivo experiments demonstrated that BSO significantly increased PRIMA-1Met efficiency.
In summary, PRIMA-1Met, alone or in combination with BSO, is potent against myeloma cells in vitro and in vivo, independent of p53 expression and TP53 status.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Tumorothèque Institut Régional du Cancer Nantes-Atlantique (Centre Hospitalo-Universitaire and Institut de Cancérologie de l'Ouest, Nantes, France) for providing us with purified myeloma cells.
B.T. was supported by a grant from Institut National du Cancer. This work was supported by a grant from Fondation Françaisepour la Recherche Contre le Myélome et les Gammapathies Monoclonales.
Authorship
Contribution: B.T., G.D., and S.M. performed experiments and participated in the design of the study; L.L. and C.G. provided purified myeloma samples and performed chromosomal analyses; S.M.-L. and T.O. performed in vivo experiments; P.M. and S.L.G. provided myeloma primary samples, participated in the design of the study, and reviewed the paper; M.A. participated in the design of the study and reviewed the paper; and C.P.-D. designed the study and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine Pellat-Deceunynck, INSERM, UMR 892, Centre de Recherches en Cancérologie Nantes Angers, IRS-UN, 8, quai Moncousu, Nantes, BP70721 F-44007 France; e-mail: catherine.pellat-deceunynck@inserm.fr.
References
Author notes
B.T. and G.D. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal