Key Points
Global lncRNA discovery reveals novel erythroid-specific lncRNAs that are dynamically expressed and targeted by GATA1, TAL1, and KLF1.
Multiple types of lncRNAs promote red cell maturation by regulating neighboring loci, including DLEU2 and a novel Band 3 enhancer lncRNA.
Abstract
Erythropoiesis is regulated at multiple levels to ensure the proper generation of mature red cells under multiple physiological conditions. To probe the contribution of long noncoding RNAs (lncRNAs) to this process, we examined >1 billion RNA-seq reads of polyadenylated and nonpolyadenylated RNA from differentiating mouse fetal liver red blood cells and identified 655 lncRNA genes including not only intergenic, antisense, and intronic but also pseudogene and enhancer loci. More than 100 of these genes are previously unrecognized and highly erythroid specific. By integrating genome-wide surveys of chromatin states, transcription factor occupancy, and tissue expression patterns, we identify multiple lncRNAs that are dynamically expressed during erythropoiesis, show epigenetic regulation, and are targeted by key erythroid transcription factors GATA1, TAL1, or KLF1. We focus on 12 such candidates and find that they are nuclear-localized and exhibit complex developmental expression patterns. Depleting them severely impaired erythrocyte maturation, inhibiting cell size reduction and subsequent enucleation. One of them, alncRNA-EC7, is transcribed from an enhancer and is specifically needed for activation of the neighboring gene encoding BAND 3. Our study provides an annotated catalog of erythroid lncRNAs, readily available through an online resource, and shows that diverse types of lncRNAs participate in the regulatory circuitry underlying erythropoiesis.
Introduction
Red blood cell development is a highly coordinated process essential throughout the lifetime of all mammals. It involves the generation of mature definitive erythrocytes from multipotent stem cells residing in the fetal liver or the adult bone marrow via cell lineage specification, proliferation, and differentiation.1 Coordination of this process requires dynamic and precise gene expression control, and disruption of erythroid regulatory networks leads to disease.2,3 Identifying novel modulators of erythrocyte production is thus crucial for finding new opportunities for treatment of erythroid disorders.
Long noncoding RNAs (lncRNAs) are transcripts longer than 200 nucleotides without functional protein-coding capacity. Large-scale studies indicate that these RNAs are pervasively transcribed in mammalian cells.4-6 Based on their genomic region of origin, lncRNAs can be classified as intergenic (lincRNAs), antisense to other genes (alncRNAs), intron-overlapping with protein-coding genes (ilncRNAs), small RNA (sRNA) hosts (shlncRNAs), enhancer-derived (elncRNAs), or pseudogene-derived (plncRNAs). Efforts to characterize lncRNAs thus far have largely focused on lincRNAs, given that as nonoverlapping transcriptional units they are readily identifiable and experimentally tractable.7 Globally, lincRNAs are expressed at lower levels but in a more cell type–specific manner than messenger RNAs (mRNAs),8 suggesting roles in lineage-specific development or in specialized cellular functions. Indeed, several lincRNAs have been implicated in modulating mammalian cell differentiation.9 The relative contributions of the full panorama of lncRNA classes to the same developmental process, however, remain poorly understood.
Here, we comprehensively characterize the landscape of lncRNAs expressed during red blood cell development in vivo. We use RNA-seq to survey the poly(A)+ and poly(A)− RNA transcriptomes of differentiating E14.5 mouse fetal liver erythroid cells and identify 655 lncRNAs of various classes, including 132 previously unannotated loci with erythroid-restricted expression. We uncover ∼100 lncRNAs with dynamic expression and chromatin patterns during differentiation, many of which are targeted by key erythroid transcription factors (TFs) GATA1, TAL1, or KLF1. These include novel erythroid-specific lncRNAs found in the nucleus that show striking patterns of developmental stage specificity and are often conserved in human. Depleting 12 such candidates with short hairpin RNAs (shRNAs) revealed critical roles in the transition from terminally differentiated erythroblasts to mature enucleated erythrocytes. One of them is derived from an enhancer region and is needed for activating the neighboring gene encoding BAND 3, the major anion transporter of the red cell membrane. Our data and workflow provide a roadmap for the identification of lncRNAs with roles in erythropoiesis, which can be readily implemented through an online resource (http://lodishlab.wi.mit.edu/data/lncRNAs/). Overall, our study provides a comprehensive catalog of erythroid lncRNAs and reveals several novel modulators of erythropoiesis.
Methods
Cell isolation, culture, and terminal differentiation assays
RNA-seq and analysis
Total RNA was isolated from mouse fetal liver TER119+ or TER119− cells using the QIAGEN miRNeasy Kit. Ribosomal RNA was depleted using the Epicentre Ribo-Zero Gold Kit. Strand-specific sequencing libraries were prepared as described12 from the total, poly(A)+ or poly(A)− RNA fractions and sequenced using the Illumina HiSeq2000 platform. Paired-end RNA-seq reads were mapped to the mouse genome (mm9 version) using TopHat,13 and transcripts were assembled de novo using Cufflinks.14 We also examined poly(A)+ RNA-seq reads from purified burst-forming unit erythroid (BFU-E) progenitors, colony-forming unit erythroid (CFU-E), and TER119+ cells15 and from 30 cell and tissue types from the mouse ENCODE consortium16 (supplemental Table 3, available on the Blood Web site). Gene-level expression for all datasets was quantified as Fragments per Kilobase of exon per Million fragments mapped (FPKM) using Cufflinks based on our de novo gene models. Differential gene expression was determined using DESeq,17 with a false discovery rate (FDR) threshold of 5%. Further details can be found in supplemental Methods. RNA-seq data from this study have been deposited at the National Center for Biotechnology Information Gene Expression Omnibus (accession number GSE52126).
lncRNA identification and classification
Once a de novo transcriptome from TER119+ and TER119− cells was assembled, to identify reliable lncRNA models, we considered only multiexonic transcripts and ran them through the following filters: (1) size selection, (2) empirical read coverage threshold, (3) known protein domain filter, (4) predicted coding potential threshold, and (5) overlap with known mRNA exon annotations filter (see supplemental Methods for specific details). To assign lncRNAs to specific classes, we examined their overlap with annotated genes from the Ensembl,18 RefSeq,19 and University of California, Santa Cruz (UCSC) Genome Browser20 databases or with enhancers annotated in E14.5 fetal liver cells.21
Single-molecule RNA fluorescence in situ hybridization and analysis
Retroviral transduction
Purified erythroid progenitors were transduced by murine stem cell virus-based retroviruses following previously described protocols.11
Flow cytometry and analysis
For all flow cytometry experiments, we gated on transduced cells (green fluorescent protein-positive subpopulation), for phenotypic analysis. The procedures for immunostaining and flow cytometry analysis of erythroid differentiation and enucleation were described previously.11,24 Average cell size was quantified as the mean of the distribution of forward scatter pulse area measurements.
Results
Global discovery of lncRNAs expressed in fetal liver and erythroid cells
The mouse fetal liver is the primary site of erythropoiesis between embryonic days 12 and 16. To catalog lncRNAs expressed during fetal erythropoiesis in vivo, we used high-throughput sequencing to survey the long RNA transcriptome of E14.5 fetal liver cells and characterized that of the erythroid lineage subpopulation. In brief, we used previously developed methods10,11 to purify fetal liver TER119-positive cells, representing a pure population of differentiating hemoglobinizing erythroblasts, and TER119-negative cells, enriched for erythroid progenitors (∼90%) but also containing cells from other hematopoietic lineages and niche cells.10 We generated strand-specific, paired-end 100-bp RNA-seq reads of total RNA depleted of rRNA from both cell populations and of poly(A)+ and poly(A)− RNA from TER119+ cells. In addition, we examined RNA-seq reads of poly(A)+ RNA from fluorescence-activated cell sorter (FACS)-purified fetal liver BFU-E progenitors, CFU-E progenitors, and TER119+ erythroblasts.15 Using TopHat,13 we mapped in total >1 billion RNA-seq reads to the mouse genome (supplemental Table 1), thus surveying the fetal erythroid differentiation transcriptome at unprecedented resolution. Transcripts were reconstructed de novo from these data using Cufflinks14 and compared with Ensembl,18 RefSeq,19 and UCSC20 gene annotations.
To identify lncRNAs with high confidence, we considered only multiexonic transcripts and discarded any that overlap with known mRNA exons in the same strand or that have predicted coding potential based on multiple orthogonal approaches (supplemental Methods). Our stringent strategy, summarized in Figure 1A, yielded 800 lncRNAs from 655 loci, all of which have no predicted functional coding capacity (supplemental Figure 1A-B). In total, our transcriptome from TER119+ and TER119− cells contained 9512 known mRNA genes, 655 lncRNA genes, and 209 genes that have unclear coding capacity based on our criteria and were thus discarded from further analysis (supplemental Figure 1C). Importantly, we identified 194 lncRNAs from 132 loci that were previously unannotated (Figure 1B), likely missed by previous databases due to their erythroid-specific expression (see below).
Identification of lncRNAs expressed in fetal liver and erythroid cells. (A) Workflow for lncRNA discovery. See text and supplemental “Methods” for details. (B) Overlap between lncRNAs annotated in Ensembl, UCSC, or RefSeq databases and lncRNAs identified in this study. (C) Definitions of different classes of lncRNAs based on their genomic region of origin. (D) Distribution of 655 lncRNAs expressed in fetal liver into different lncRNA classes.
Identification of lncRNAs expressed in fetal liver and erythroid cells. (A) Workflow for lncRNA discovery. See text and supplemental “Methods” for details. (B) Overlap between lncRNAs annotated in Ensembl, UCSC, or RefSeq databases and lncRNAs identified in this study. (C) Definitions of different classes of lncRNAs based on their genomic region of origin. (D) Distribution of 655 lncRNAs expressed in fetal liver into different lncRNA classes.
To classify the repertoire of fetal liver lncRNAs, we examined their overlap with annotated genes18-20 or enhancers21 (Figure 1C) and systematically assigned them to lncRNA classes (supplemental Figure 1D; supplemental Methods). Our strategy identified 299 lincRNAs, 153 alncRNAs, 92 ilncRNAs, 52 elncRNAs, 27 shlncRNAs, and 3 plncRNAs (Figure 1D; supplemental Table 2), as well as 29 lncRNAs that could not be classified. The majority of our intergenic, antisense, intronic, sRNA-hosting, and pseudogene lncRNAs that are found in Ensembl are annotated as such (supplemental Figure 1F), thus validating our strategy. For enhancer lncRNAs, as expected,25,26 we found enrichment around the transcriptions start site (TSS) for H3K27Ac and H3K4me1 over H3K4me3, as well as for serine 5 phosphorylated RNA Pol II, in E14.5 fetal liver cells (supplemental Figure 1E). We further noted that our lncRNAs share similar features with those described in previous global studies in mice and humans,8,27-30 in terms of structural, expression, and conservation properties (supplemental Text 1 and 2; supplemental Figures 2 and 3).
High tissue specificity among fetal liver and erythroid lncRNAs
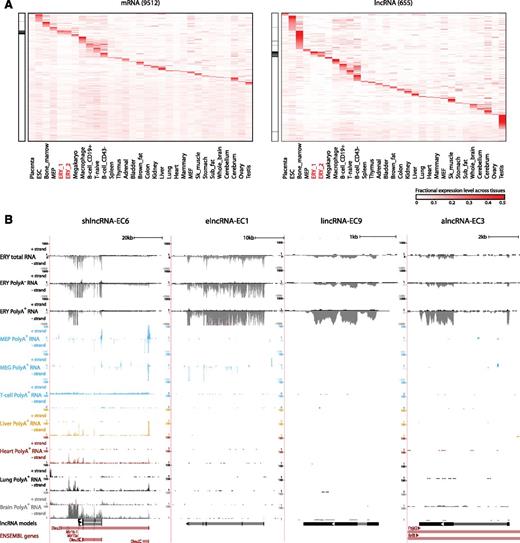
Our high-resolution transcriptomic survey of TER119+ and TER119− cells covers genes from erythroid cells and from minor cell populations of other lineages. Moreover, genes expressed at basal levels in fetal liver cells may be more characteristic of other tissues. To globally examine tissue specificity, we quantified the expression of fetal liver-expressed genes across a compendium of 30 primary cell and tissue types purified, sequenced, and analyzed using common guidelines for the mouse ENCODE consortium16 (Figure 2; supplemental Table 3). For each gene, we scored the specificity of its expression in a given tissue as the fraction of the total expression across tissues that it represents, equivalent to its fractional expression level. Based on these scores, we find exquisite patterns of tissue-specific enrichment for both mRNAs and lncRNAs, including genes highly specific to each of the hematopoietic lineages examined (Figure 2A). As expected,8,30 lncRNAs show greater tissue specificity than mRNAs (P < 10−15, Kolmogorov-Smirnov test), which holds true across lncRNA classes and matched expression ranges (supplemental Figure 4A-B). However, there were notable differences in tissue specificity patterns among lncRNA families (supplemental Figure 4A,C). For example, elncRNAs are the only class enriched in both fetal erythroblasts and in the adult bone marrow, suggesting that they originate from enhancers active at both fetal and adult stages.
Tissue specificity of fetal liver and erythroid lncRNAs. (A) Relative abundance of mRNA and lncRNA genes (rows) expressed in fetal liver across 30 primary cell and tissue types from the mouse ENCODE consortium (columns). Color intensity represents the fractional gene-level expression across all tissues examined. ERY_1 and ERY_2 (red) are fetal liver TER119+ erythroblast replicates. Tissue expression was quantified based on gene models from our de novo assembly using Cufflinks. Black bars in the left panels highlight empirically defined erythroid-restricted genes. (B) Examples of erythroid-enriched lncRNA loci. These loci were selected based on their expression, regulation, and tissue specificity features (see text). Images from the UCSC Genome Browser depict RNA-seq signal as the density of mapped strand-specific RNA-seq reads. The plus strand (transcribed left to right) and minus strand (transcribed right to left) are denoted to the left of the tracks. Tracks 1 to 6 show in black the RNA-seq signal of total, poly(A)−, or poly(A)+ RNA from fetal liver TER119+ erythroblasts (ERY). Tracks 7 to 12 depict in light blue the RNA-seq signal of poly(A)+ RNA from other hematopoietic cells: adult megakaryocyte-erythroid progenitors (MEP), fetal megakaryocytes (MEG), and adult T-naïve cells (T-cell), all from the ENCODE consortium. Tracks 13 to 20 show the RNA-seq signal of poly(A)+ RNA from other tissues from the ENCODE consortium: adult liver (yellow), adult heart (red), adult lung (black), and E14.5 whole brain (gray). The bottom tracks depict lncRNA transcript models inferred by de novo assembly using Cufflinks (black) and Ensembl gene annotations (red). Left-to-right arrows indicate transcripts in the plus strand; right-to-left arrows indicate transcripts in the minus strand. Note that all lncRNA transcripts shown are transcribed in the minus strand.
Tissue specificity of fetal liver and erythroid lncRNAs. (A) Relative abundance of mRNA and lncRNA genes (rows) expressed in fetal liver across 30 primary cell and tissue types from the mouse ENCODE consortium (columns). Color intensity represents the fractional gene-level expression across all tissues examined. ERY_1 and ERY_2 (red) are fetal liver TER119+ erythroblast replicates. Tissue expression was quantified based on gene models from our de novo assembly using Cufflinks. Black bars in the left panels highlight empirically defined erythroid-restricted genes. (B) Examples of erythroid-enriched lncRNA loci. These loci were selected based on their expression, regulation, and tissue specificity features (see text). Images from the UCSC Genome Browser depict RNA-seq signal as the density of mapped strand-specific RNA-seq reads. The plus strand (transcribed left to right) and minus strand (transcribed right to left) are denoted to the left of the tracks. Tracks 1 to 6 show in black the RNA-seq signal of total, poly(A)−, or poly(A)+ RNA from fetal liver TER119+ erythroblasts (ERY). Tracks 7 to 12 depict in light blue the RNA-seq signal of poly(A)+ RNA from other hematopoietic cells: adult megakaryocyte-erythroid progenitors (MEP), fetal megakaryocytes (MEG), and adult T-naïve cells (T-cell), all from the ENCODE consortium. Tracks 13 to 20 show the RNA-seq signal of poly(A)+ RNA from other tissues from the ENCODE consortium: adult liver (yellow), adult heart (red), adult lung (black), and E14.5 whole brain (gray). The bottom tracks depict lncRNA transcript models inferred by de novo assembly using Cufflinks (black) and Ensembl gene annotations (red). Left-to-right arrows indicate transcripts in the plus strand; right-to-left arrows indicate transcripts in the minus strand. Note that all lncRNA transcripts shown are transcribed in the minus strand.
We next focused on the subset of genes showing erythroid-specific expression, which we defined as tissue-restricted genes having fetal erythroblasts as the tissue with the maximal tissue specificity score (supplemental Methods; highlighted in Figure 2A). These genes comprised 7% and 8% of fetal liver-expressed mRNAs and lncRNAs, respectively. Previously unannotated lncRNAs are enriched in this group relative to annotated ones (supplemental Figure 4D), highlighting the importance of our focus on erythroid cells. In contrast, broadly expressed lncRNAs are mostly depleted or expressed at background levels in fetal erythroblasts (supplemental Figure 4E). By way of example, shown in Figure 2B are four lncRNAs, shlncRNA-EC6, elncRNA-EC1, lincRNA-EC9, and alncRNA-EC3, that we focus on later because of their erythroid specificity, promoter targeting by erythroid TFs, high expression in erythroblasts, and induction during terminal differentiation (see below). elncRNA-EC1, lincRNA-EC9, and alncRNA-EC3 are expressed in erythroblasts but not in the closely related megakaryocyte or megakaryocyte-erythroid progenitor or in other tissues examined (Figure 2B). In contrast, the shlncRNA-EC6 locus, encoding the known lncRNA DLEU2, is broadly expressed, but it is transcribed from a different promoter in erythroblasts vs other cell types, including closely related lineages (Figure 2B). Interestingly, processing of this erythroid-restricted isoform, presumably to release microRNAs 16-1 and 15a from the poly(A)+ precursor, generates mature poly(A)+ and poly(A)− transcripts of similar stability, as evidenced by comparable levels in their respective RNA fractions (supplemental Figure 9A).
lncRNAs are dynamically regulated during erythropoiesis
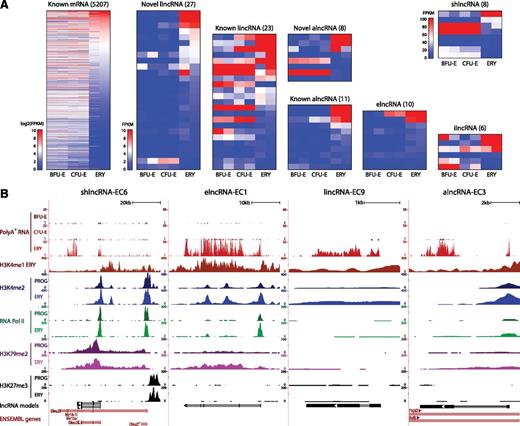
To examine the regulation of lncRNAs during erythropoiesis, we focused on the subset of fetal liver-expressed genes that are reliably detected in FACS-purified BFU-Es, CFU-Es, or TER119+ erythroblasts. We considered only genes expressed in both replicates of ≥1 stage from BFU-Es to erythroblasts, resulting in 275 erythroid-expressed lncRNAs (supplemental Table 4), and then used DESeq17 to identify differentially expressed ones (P < .05, DESeq test). We identified 728 differentially expressed mRNAs increasing more than twofold between progenitors and erythroblasts that as expected encode proteins enriched for erythroid-specific roles (supplemental Figure 5A), thus validating our approach. We also identified 96 lncRNAs of all classes that are differentially expressed during erythropoiesis (Figure 3A). These comprised mostly lncRNAs that become strongly induced as progenitors differentiate into erythroblasts, as exemplified by the 4 lncRNAs examined in Figure 2B (Figure 3B). Importantly, we find that lncRNAs exhibit greater expression variability than mRNAs through the stages of differentiation, with novel erythroid-enriched lncRNAs being more dynamically expressed than previously annotated ones (supplemental Figure 5B-C). Moreover, we confirmed that, as with mRNAs,20 changes in lncRNA expression were reflected at the chromatin level (supplemental Text 3; supplemental Figures 6-7). The most predictive activation and elongation histone marks were H3K4me2 and H3K79me2, respectively (Pearson’s r = 0.71, P < 10−12 and r = 0.46, P < 10−4, Fisher’s exact test), shown for the highly induced lncRNAs in Figure 3B. By contrast, repressive H3K27me3 marking was generally depleted near active TSSs, as exemplified at the shlncRNA-EC6 locus, where the repressed, distal promoter is marked by H3K27me3. Notably, H3K27me3 marking of this promoter is already established at the committed progenitor stage (Figure 3B).
Dynamic expression patterns of lncRNAs during erythroid differentiation. (A) Abundance of mRNAs and lncRNAs that are differentially expressed during erythropoiesis, as determined by DESeq at a 5% false discovery threshold. Shown are absolute gene expression estimates (FPKM) from poly(A)+ RNA-seq of FACS-purified BFU-Es, CFU-Es, and TER119+ erythroblasts (ERY) (2 replicates each), based on gene models from our de novo assembly using Cufflinks. (B) Examples of differentially expressed lncRNA loci, the same RNAs as in Figure 2B. Images from the UCSC Genome Browser depict RNA-seq signal as the density of mapped RNA-seq reads and chromatin immunoprecipitation sequencing (ChIP-seq) signal as the density of processed signal enrichment. Tracks 1 to 3 show in red the non–strand-specific RNA-seq signal of poly(A)+ RNA from FACS-purified fetal liver BFU-Es, CFU-Es, and TER119+ erythroblasts (ERY). Tracks 4 to 12 depict the ChIP-seq signal for H3K4me1, a chromatin mark enriched in promoter and enhancer regions, in ERY (dark red); H3K4me2, associated with transcriptional activation, in erythroid progenitor-enriched fetal liver cells (PROG) and ERY (dark and light blue); serine 5 phosphorylated RNA Pol II, enriched at the TSS of active genes, in PROG and ERY (dark and light green); H3K79me2, associated with transcriptional elongation, in PROG and ERY (dark and light purple); and H3K27me3, associated with transcriptional repression, in PROG and ERY (black). The bottom tracks depict lncRNA transcript models and Ensembl gene annotations as in Figure 2B.
Dynamic expression patterns of lncRNAs during erythroid differentiation. (A) Abundance of mRNAs and lncRNAs that are differentially expressed during erythropoiesis, as determined by DESeq at a 5% false discovery threshold. Shown are absolute gene expression estimates (FPKM) from poly(A)+ RNA-seq of FACS-purified BFU-Es, CFU-Es, and TER119+ erythroblasts (ERY) (2 replicates each), based on gene models from our de novo assembly using Cufflinks. (B) Examples of differentially expressed lncRNA loci, the same RNAs as in Figure 2B. Images from the UCSC Genome Browser depict RNA-seq signal as the density of mapped RNA-seq reads and chromatin immunoprecipitation sequencing (ChIP-seq) signal as the density of processed signal enrichment. Tracks 1 to 3 show in red the non–strand-specific RNA-seq signal of poly(A)+ RNA from FACS-purified fetal liver BFU-Es, CFU-Es, and TER119+ erythroblasts (ERY). Tracks 4 to 12 depict the ChIP-seq signal for H3K4me1, a chromatin mark enriched in promoter and enhancer regions, in ERY (dark red); H3K4me2, associated with transcriptional activation, in erythroid progenitor-enriched fetal liver cells (PROG) and ERY (dark and light blue); serine 5 phosphorylated RNA Pol II, enriched at the TSS of active genes, in PROG and ERY (dark and light green); H3K79me2, associated with transcriptional elongation, in PROG and ERY (dark and light purple); and H3K27me3, associated with transcriptional repression, in PROG and ERY (black). The bottom tracks depict lncRNA transcript models and Ensembl gene annotations as in Figure 2B.
lncRNAs are targets of core erythroid transcription factors
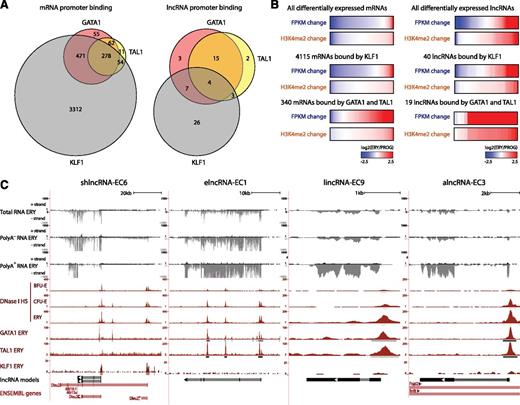
We reasoned that if differentially expressed lncRNAs play roles during erythropoiesis, they should be targeted by erythroid-important TFs. To test this, we examined genome-wide maps of GATA1, TAL1, and KLF1 occupancy determined by ChIP-seq in fetal liver TER119+ erythroblasts.31,32 Binding sites for these factors, inferred by model-based analysis of ChIP-seq (MACS)33 (empirical FDR <0.05), were intersected with the promoter-proximal regions (TSS ± 1 kb) of differentially expressed lncRNAs or mRNAs. As expected,34 all 3 factors co-occupied the promoters of 278 mRNA genes differentially expressed during erythropoiesis, including 136 that are up-regulated more than twofold and encode proteins with erythroid-specific roles (Figure 4A; supplemental Figure 8A), thus validating our approach. We then found that 60 of 96 differentially expressed lncRNAs are indeed bound at their promoters by GATA, TAL1, or KLF1 in erythroblasts (Figure 4A). We also found that promoter-proximal co-occupancy by GATA1 and TAL1 for mRNAs and lncRNAs is significantly associated with gene induction (P < 10−15 and P < 10−9, respectively, Wilcoxon test) and promoter H3K4me2 marking (P < 10−15 and P < 10−4, respectively, Kolmogorov-Smirnov test) (Figure 4B; supplemental Figure 8B), extending previous observations of mRNAs.32,34,35 In contrast, proximal binding by KLF1 alone seems to be a poor predictor of gene expression change (Figure 4B). Binding peaks for GATA1, TAL1, and KLF1 are shown for our lncRNA models in Figure 4C. The coincidence of these peaks with DNase I hypersensitive sites (Figure 4C) and with RNA pol II binding and active chromatin marks (Figure 3B) in these genes strongly supports their specific regulation by these factors. Importantly, both TF binding events and chromatin architecture are conserved in the human K562 erythroleukemia cell line for the DLEU2 human ortholog and for the putative ortholog of elncRNA-EC1, inferred by local alignment and synteny (supplemental Figure 8D; supplemental Methods). For these 2 genes, we obtained direct evidence of regulation by GATA1 by confirming their time-dependent activation after GATA1 restoration in the mouse G1E-ER4 cell line36,37 (supplemental Figure 8C).
lncRNAs are targeted by core erythroid transcription factors. (A) Binding of GATA1, TAL1, and KLF1 transcription factors within promoter-proximal regions (TSS ± 1 kb) of (left) mRNAs and (right) lncRNAs that are differentially expressed during erythropoiesis (see text). (B) Changes in expression and promoter-proximal (TSS ± 1 kb) H3K4me2 levels for all differentially expressed mRNA or lncRNA genes, for the subset of genes bound by KLF1 or for those bound by both GATA and TAL1. Changes are shown as the log2 ratio of the levels in TER119+ erythroblasts (ERY) to the levels in erythroid progenitor-enriched fetal liver cells (PROG). (C) Examples of differentially expressed lncRNA loci that are bound proximally by GATA1, TAL1, or KLF1, the same RNAs as in Figures 2B and 3B. Images from the UCSC Genome Browser depict the RNA-seq signal as the density of mapped RNA-seq reads, DNase I hypersensitivity (HS) signal as the density of mapped sequencing tags, and ChIP-seq signal as the density of processed signal enrichment. Tracks 1 to 6 show in black the strand-specific RNA-seq signal in the plus strand or minus strand (denoted to the left of the tracks) of total, poly(A)−, or poly(A)+ RNA from fetal liver TER119+ erythroblasts (ERY). Tracks 7 to 9 depict in red the signal for DNase I HS, associated with open chromatin, in BFU-Es, CFU-Es, and ERY. Tracks 10 to 12 show in red the ChIP-seq signal for GATA1, TAL1, and KLF1, respectively, in ERY. Peaks of signal enrichment are shown in gray under the DNase I HS tracks (determined by I-max, empirical FDR <1%) and under the GATA1, TAL1, and KLF1 tracks (determined by MACS, empirical FDR <5%). The bottom tracks depict lncRNA transcript models and Ensembl gene annotations as in Figure 2B.
lncRNAs are targeted by core erythroid transcription factors. (A) Binding of GATA1, TAL1, and KLF1 transcription factors within promoter-proximal regions (TSS ± 1 kb) of (left) mRNAs and (right) lncRNAs that are differentially expressed during erythropoiesis (see text). (B) Changes in expression and promoter-proximal (TSS ± 1 kb) H3K4me2 levels for all differentially expressed mRNA or lncRNA genes, for the subset of genes bound by KLF1 or for those bound by both GATA and TAL1. Changes are shown as the log2 ratio of the levels in TER119+ erythroblasts (ERY) to the levels in erythroid progenitor-enriched fetal liver cells (PROG). (C) Examples of differentially expressed lncRNA loci that are bound proximally by GATA1, TAL1, or KLF1, the same RNAs as in Figures 2B and 3B. Images from the UCSC Genome Browser depict the RNA-seq signal as the density of mapped RNA-seq reads, DNase I hypersensitivity (HS) signal as the density of mapped sequencing tags, and ChIP-seq signal as the density of processed signal enrichment. Tracks 1 to 6 show in black the strand-specific RNA-seq signal in the plus strand or minus strand (denoted to the left of the tracks) of total, poly(A)−, or poly(A)+ RNA from fetal liver TER119+ erythroblasts (ERY). Tracks 7 to 9 depict in red the signal for DNase I HS, associated with open chromatin, in BFU-Es, CFU-Es, and ERY. Tracks 10 to 12 show in red the ChIP-seq signal for GATA1, TAL1, and KLF1, respectively, in ERY. Peaks of signal enrichment are shown in gray under the DNase I HS tracks (determined by I-max, empirical FDR <1%) and under the GATA1, TAL1, and KLF1 tracks (determined by MACS, empirical FDR <5%). The bottom tracks depict lncRNA transcript models and Ensembl gene annotations as in Figure 2B.
Validation of candidate lncRNAs reveals nuclear localization and complex developmental patterns
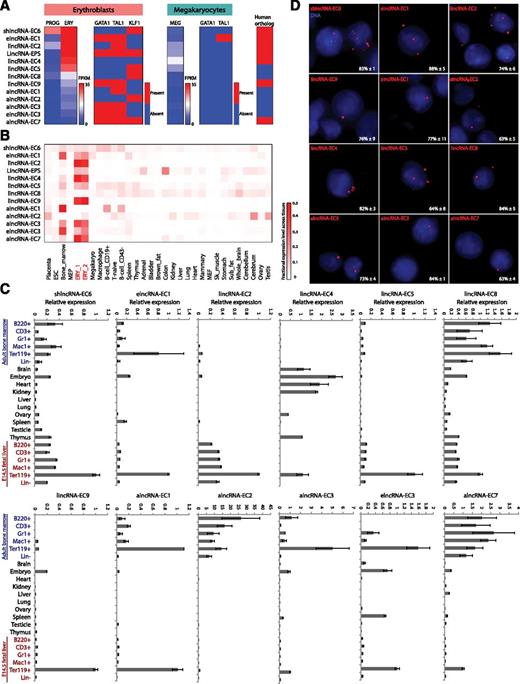
To select candidates for functional studies, we devised a strategy that integrates the experimental and computational analyses described above to stringently identify lncRNAs likely to play roles in erythropoiesis (supplemental Methods). Briefly, we focused on differentially expressed lncRNAs with independent evidence of active transcription. Next, we required that they be targets of GATA1, TAL1, or KLF1 or that their expression be erythroid specific. Finally, we ranked them first by their relative fold increase in expression between progenitors and erythroblasts and then by their absolute expression in erythroblasts. This strategy yielded 6 lincRNAs, 4 alncRNAs, 2 elncRNAs, and 1 shlncRNA as the top candidate modulators of erythropoiesis. The expression, regulation, and conservation features of these candidates are summarized in Figure 5A-B and are shown in detail in supplemental Figure 9. Ranked fourth among them is LincRNA-EPS, which we previously found to promote erythroid differentiation by preventing apoptosis,38 thus validating our approach. Several of the new candidates are previously unannotated lncRNAs that are induced to similar levels as LincRNA-EPS during terminal differentiation. These include erythroid-specific ones that are cotargeted by GATA1 and TAL1 (Figure 4C; supplemental Figure 9B,G,J), but also more broadly expressed ones that are targeted by KLF1 instead (supplemental Figure 9E-F,I). We note that lincRNA-EC4, not targeted proximally by any of these factors, has been independently characterized recently (Lincred139 ) and shown to be targeted distally by KLF1. Overall, the expression and regulation features of our candidates in erythroblasts are generally absent from megakaryocytes16 (Figure 5A), suggesting erythroid-specific functions.
Selection and validation of lncRNA targets. (A) Summary of expression, regulation, and conservation features of the top candidate lncRNA modulators of erythropoiesis (see text). Expression: shown are absolute gene expression estimates (FPKM) from RNA-seq of total RNA from erythroid progenitor-enriched fetal liver cells (PROG) and TER119+ erythroblasts (ERY) or of poly(A)+ RNA from primary megakaryocytes (MEG), quantified as in Figure 3. Regulation: heatmaps represent whether promoter-proximal binding by GATA1, TAL1, of KLF1, analyzed as in Figure 4, is seen in ERY or MEG. Conservation: heatmap represents whether an orthologous region, identified by local alignment and synteny, is found in the human genome (see supplemental Methods for details). (B) Relative abundance of the top lncRNA candidates across 30 mouse primary tissue and cell types from ENCODE, determined as in Figure 2. Color intensity represents the fractional expression level across all tissues examined. ERY_1 and ERY_2 (red) are TER119+ fetal liver erythroblast biological replicate experiments. (C) Relative expression of the top lncRNA candidates across mouse organs and cells of different tissues and developmental stages, as determined by qPCR. Expression levels were normalized to those of 18S rRNA, and fold changes were calculated relative to fetal TER119+ erythroblast levels. Data are shown as mean ± standard error of the mean (SEM; n = 3). (D) Detection of individual lncRNA transcripts by single-molecule RNA FISH. Shown are maximum z-stack projections of fluoresce microscopy images of fixed TER119+ erythroblasts hybridized to singly-labeled RNA FISH probes. lncRNA molecules are pseudocolored red and DAPI-stained nuclei are pseudocolored blue. For each panel, the mean ± SEM (n = 2) percent of nuclear-localized transcripts is shown at the bottom right corner.
Selection and validation of lncRNA targets. (A) Summary of expression, regulation, and conservation features of the top candidate lncRNA modulators of erythropoiesis (see text). Expression: shown are absolute gene expression estimates (FPKM) from RNA-seq of total RNA from erythroid progenitor-enriched fetal liver cells (PROG) and TER119+ erythroblasts (ERY) or of poly(A)+ RNA from primary megakaryocytes (MEG), quantified as in Figure 3. Regulation: heatmaps represent whether promoter-proximal binding by GATA1, TAL1, of KLF1, analyzed as in Figure 4, is seen in ERY or MEG. Conservation: heatmap represents whether an orthologous region, identified by local alignment and synteny, is found in the human genome (see supplemental Methods for details). (B) Relative abundance of the top lncRNA candidates across 30 mouse primary tissue and cell types from ENCODE, determined as in Figure 2. Color intensity represents the fractional expression level across all tissues examined. ERY_1 and ERY_2 (red) are TER119+ fetal liver erythroblast biological replicate experiments. (C) Relative expression of the top lncRNA candidates across mouse organs and cells of different tissues and developmental stages, as determined by qPCR. Expression levels were normalized to those of 18S rRNA, and fold changes were calculated relative to fetal TER119+ erythroblast levels. Data are shown as mean ± standard error of the mean (SEM; n = 3). (D) Detection of individual lncRNA transcripts by single-molecule RNA FISH. Shown are maximum z-stack projections of fluoresce microscopy images of fixed TER119+ erythroblasts hybridized to singly-labeled RNA FISH probes. lncRNA molecules are pseudocolored red and DAPI-stained nuclei are pseudocolored blue. For each panel, the mean ± SEM (n = 2) percent of nuclear-localized transcripts is shown at the bottom right corner.
To validate our candidates, we used quantitative real-time polymerase chain reaction (qPCR) to measure expression across hematopoietic lineages purified from fetal liver or adult bone marrow (Figure 5C). Consistent with our RNA-seq data, we confirmed hematopoietic tissue specificity for all of them except for lincRNA-EC4 (also enriched in brain, heart, and kidney). Strikingly, most of our lincRNAs are enriched in fetal liver but not adult erythroblasts (Figure 5C). By contrast, alncRNA-EC3 and the 2 elncRNAs examined are enriched in both fetal and adult erythroblasts. A third pattern was apparent for shlncRNA-EC6, lincRNA-EC8, and the remaining 3 alncRNAs, which are expressed during both fetal and adult hematopoiesis but are only erythroid enriched at the fetal stage. Thus, lncRNA expression can be highly specific to developmental stage, even within the same cell lineage.
We next used single-molecule RNA FISH (smFISH)22 to visualize lncRNA transcripts in FACS-purified fetal liver TER119+ erythroblasts (Figure 5D). These experiments revealed our candidates to be predominantly nuclear, which we confirmed by cellular fractionation followed by qPCR (supplemental Figure 10). smFISH also indicated mean lncRNA levels of ∼1 to 10 transcripts per cell, consistent with previous measurements in mouse and human.40,41
lncRNAs of multiple classes regulate red cell maturation
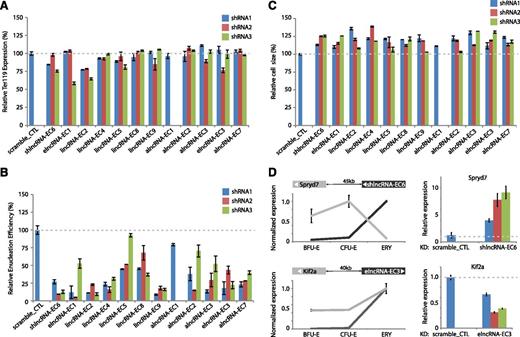
To conduct loss-of-function experiments, we generated 3 shRNAs for each lncRNA with the exception of alncRNA-EC1, where only 1 shRNA was possible due to extensive repeats. For alncRNAs, shRNAs were designed to target regions that either do not overlap the transcript on the opposite strand or only overlap its introns. We introduced each shRNA via retroviral transduction into lineage-negative fetal liver cells, which are enriched for erythroid progenitors,15 and then cultured them in erythropoietin-containing media to induce ex vivo terminal proliferation and differentiation.10 We confirmed efficient knockdown of all candidates (mean 27.5-99.3% knockdown per shRNA; supplemental Figure 11). Flow cytometric analysis was then performed to evaluate 3 hallmarks of erythropoiesis: expression of the TER119 marker, cell size reduction, and subsequent enucleation.24 Knockdown of each lncRNA candidate severely impaired enucleation, as evidenced by a mean 20% to 85% reduction in enucleation efficiency relative to scrambled shRNA, reproducible across separate shRNAs (Figure 6B; all P < 10−16, Student t-test). Cells inhibited for these lncRNAs did induce TER119, an early event during terminal differentiation (Figure 6A), but exhibited greater cell size relative to control (11-26% greater average cell size, all P < .05, Student t-test; Figure 6C), consistent with retention of immature but terminally differentiated erythroblasts. These results indicate that our lncRNAs exert their functions after TER119 activation but before terminal red cell maturation. The precise events regulated by the lncRNAs are the subject of ongoing research.
Modulation of red cell maturation by multiple types of lncRNAs. (A) Relative expression of the early erythroid differentiation marker TER119 in erythroid progenitor-enriched fetal liver cells transduced with retroviral vectors encoding control or lncRNA-targeting shRNAs and induced to differentiate in culture. Expression levels were determined by qPCR, normalized to those of 18S rRNA, and are shown as percentage of the levels in the control shRNA experiment (dotted gray line). Data are mean ± SEM (n = 2). (B) Relative cell size of cells treated as in A. Average cell sizes were determined by flow cytometry (Methods) and are shown as percentage of the values for the control shRNA experiment (dotted gray line). Data are mean ± SEM (n = 2). (C) Relative enucleation efficiency of cells treated as in A. Enucleation efficiency was determined by flow cytometry (Methods) and is shown as percentage of the values for the control shRNA experiment (dotted gray line). Data are shown as mean ± SEM (n = 2). (D) (Top left) SPRYD7 (light gray) is anticorrelated in expression with its neighbor shlncRNA-EC6 (dark gray) during erythropoiesis. (Top right) depletion of shlncRNA-EC6 with separate shRNAs in ex vivo–differentiated TER119+ erythroblasts results in reproducible up-regulation of SPRYD7 relative to scramble shRNA control (data are mean ± SEM; n = 3). (Bottom left) KIF2A (light gray) is coordinated in expression with neighboring elncRNA-EC3 (dark gray) during erythroid differentiation. (Bottom right) inhibiting elncRNA-EC3 with separate shRNAs leads to reduced expression of KIF2A relative to scramble shRNA control (data are mean ± SEM; n = 3).
Modulation of red cell maturation by multiple types of lncRNAs. (A) Relative expression of the early erythroid differentiation marker TER119 in erythroid progenitor-enriched fetal liver cells transduced with retroviral vectors encoding control or lncRNA-targeting shRNAs and induced to differentiate in culture. Expression levels were determined by qPCR, normalized to those of 18S rRNA, and are shown as percentage of the levels in the control shRNA experiment (dotted gray line). Data are mean ± SEM (n = 2). (B) Relative cell size of cells treated as in A. Average cell sizes were determined by flow cytometry (Methods) and are shown as percentage of the values for the control shRNA experiment (dotted gray line). Data are mean ± SEM (n = 2). (C) Relative enucleation efficiency of cells treated as in A. Enucleation efficiency was determined by flow cytometry (Methods) and is shown as percentage of the values for the control shRNA experiment (dotted gray line). Data are shown as mean ± SEM (n = 2). (D) (Top left) SPRYD7 (light gray) is anticorrelated in expression with its neighbor shlncRNA-EC6 (dark gray) during erythropoiesis. (Top right) depletion of shlncRNA-EC6 with separate shRNAs in ex vivo–differentiated TER119+ erythroblasts results in reproducible up-regulation of SPRYD7 relative to scramble shRNA control (data are mean ± SEM; n = 3). (Bottom left) KIF2A (light gray) is coordinated in expression with neighboring elncRNA-EC3 (dark gray) during erythroid differentiation. (Bottom right) inhibiting elncRNA-EC3 with separate shRNAs leads to reduced expression of KIF2A relative to scramble shRNA control (data are mean ± SEM; n = 3).
lncRNAs have been previously reported to act in cis or in trans via diverse mechanisms.9,42-44 To explore how our lncRNAs regulate erythropoiesis, we examined the expression of their closest or overlapping neighbor by qPCR following lncRNA depletion. For 9 of 12 lncRNAs, we did not observe consistent changes in the expression of their nearest neighbor (supplemental Figure 12). In contrast, inhibiting shlncRNA-EC6/DLEU2 caused up-regulation of SPRYD7/CLLD6, residing ∼45 kb away (Figure 6D). No function is known for the SPRYD7 protein, although a RNA binding role has been proposed.45 Our results suggest that DLEU2 may act in cis to repress expression of its neighbor SPRYD7, consistent with their anticorrelated expression patterns. Indeed, a link between human DLEU2 expression and the in cis epigenetic repression of its gene neighbors, including SPRYD7, has recently been proposed.46 In contrast, depleting elncRNA-EC3 leads to ∼35% to 70% loss of KIF2A expression, which resides ∼40 kb away (Figure 6D). KIF2A is a kinesin motor involved in microtubule dynamics that is required for normal mitotic progression,47,48 but no specific role during erythropoiesis has been described. elncRNA-EC3 is transcribed from an erythroid-restricted enhancer cobound at multiple sites by GATA1 and TAL1 (Figures 3B and 4C; supplemental Figure 9K), and activation of elncRNA-EC3 coincides with the more than twofold up-regulation of its neighbor KIF2A (Pearson’s r = 0.99, P < 1 × 10−2, Fisher’s exact test) in erythroblasts (Figure 6D). Thus, elncRNA-EC3 may also act in cis to enhance KIF2A expression in the erythroid lineage.
alncRNA-EC7 is an enhancer RNA needed for expression of neighboring Band 3
Close inspection of the alncRNA-EC7 locus in fetal erythroblasts reveals a 6.9-kb region of widespread H3K4me1 and H3K27Ac marking centered around a 5.2-kb region of open chromatin bound by RNA Pol II, GATA1, TAL1, and KLF1 (Figure 7A), hallmarks of a large enhancer. The site is bidirectionally transcribed into long RNAs that are spliced and polyadenylated, as indicated by our de novo transcript reconstruction from poly(A)+ RNA, and into RNAs spanning shorter regions that accumulate in the poly(A)− fraction (Figure 7A). The latter are recognized as eRNAs, short poly(A)− species that mark active enhancers.49,50 The capacity of active enhancers to also produce the former type of transcript (long ncRNAs) has only recently been appreciated,51 but their role in enhancer function has remained unclear. We find that targeting shRNAs against the enhancer-derived alncRNA-EC7 causes >80% depletion of neighboring BAND 3/SLC4A1 ∼10 kb away (Figure 7D). BAND 3 is the major anion exchanger of the erythrocyte membrane, and its mutation can lead to hemolytic anemias.52,53 Our data suggest that alncRNA-EC7 can act in cis to enhance BAND 3 expression, consistent with their coexpression across tissues (Pearson’s r = 0.92, P < 10−12, Fisher’s exact test; Figure 7C) and during erythropoiesis (Figure 7D). Moreover, smFISH revealed that alncRNA-EC7 is retained at a focal point in the nucleus (Figure 6D), suggesting that it may not diffuse beyond its transcription site. The alncRNA-EC7 enhancer site is conserved in human, as evidenced by chromatin architecture, transcriptional activity, and TF occupancy at the syntenic locus in K562 cells (Figure 7B). Importantly, mapping of long-range chromatin interactions associated with CCCTC binding factor (CTCF)54 in these cells provides direct evidence of chromatin looping between the enhancer site and the BAND 3 promoter-proximal region (Figure 7B). This is also supported by mapping of chromatin interactions associated with RNA Pol II by the same method (supplemental Figure 13). Together, these data strongly support a model whereby alncRNA-EC7 modulates erythropoiesis in part by mediating enhancer looping to activate the BAND 3 locus.
alncRNA-EC7 is an enhancer RNA needed for expression of neighboring BAND 3. (A) alncRNA-EC7 marks an enhancer site proximal to BAND 3. Images from the UCSC Genome Browser depict RNA-seq signal as the density of mapped RNA-seq reads, DNase I hypersensitivity (HS) signal as the density of mapped sequencing tags, and ChIP-seq signal as the density of processed signal enrichment. Tracks 1 to 4 show in black the strand-specific RNA-seq signal in the plus strand or minus strand (denoted to the left of the tracks) of poly(A)− or poly(A)+ RNA from fetal liver TER119+ erythroblasts (ERY). Track 5 depicts in red the signal for DNase I HS, associated with open chromatin, in ERY. Tracks 6 to 8 show in red the ChIP-seq signal for GATA1, TAL1, and KLF1, respectively, in ERY. Tracks 9 and 10 depict the ChIP-seq signal for H3K4me1, enriched along promoter and enhancer regions, in ERY (dark red), and H3K27Ac, associated with active promoters and enhancers, in fetal liver cells (yellow). Peaks of signal enrichment are shown in gray under the DNase I HS track (determined by I-max, empirical FDR <1%) and under the GATA1, TAL1, KLF1, H3K4me1, and H3K27Ac tracks (determined by MACS, empirical FDR <5%). The bottom tracks depict lncRNA transcript models inferred by de novo assembly using Cufflinks (black) and Ensembl gene annotations (red). Shown at the top are the target sites of 3 shRNAs designed against the transcripts from the enhancer site. (B) The alncRNA-EC7 locus is conserved in human. Images from the UCSC Genome Browser depict RNA-seq signal, DNase I HS signal, and ChIP-seq signal in K562 cells as in A. Track 1 shows in blue the non–strand-specific RNA-seq signal of poly(A)+ RNA. Tracks 2 and 3 depict in light blue the ChIP-seq signal for H3K4me1, enriched along promoter and enhancer regions, and H3K27Ac, associated with active promoters and enhancers. Track 4 depicts in dark blue the signal for DNase I HS, associated with open chromatin. Tracks 5 to 8 show in dark blue the ChIP-seq signal for RNA Pol II, GATA1, TAL1, and p300 (enriched at promoter and enhancer sites), respectively. Shown at the bottom are lncRNA transcript models based on our detection of orthologous genomic regions from local alignment and synteny to the mouse genome (black; supplemental Methods) and Ensembl gene annotations (red). The last track depicts in red chromatin interactions associated with binding of the CTCF binding factor, determined by ChIA-PET. (C) Relative abundance of alncRNA-EC7 and Band 3 mRNA across 30 mouse primary tissue and cell types from ENCODE, determined as in Figure 2. Color intensity represents the fractional expression level across all tissues examined. ERY_1 and ERY_2 (red) are TER119+ fetal liver erythroblast biological replicate experiments. (D) (Top) Band 3 expression is coordinated with that of neighboring alncRNA-EC7 during differentiation. (Bottom) Inhibiting alncRNA-EC7 with the shRNAs shown in A abolishes expression of BAND 3 relative to scramble shRNA control (data are mean ± SEM; n = 3).
alncRNA-EC7 is an enhancer RNA needed for expression of neighboring BAND 3. (A) alncRNA-EC7 marks an enhancer site proximal to BAND 3. Images from the UCSC Genome Browser depict RNA-seq signal as the density of mapped RNA-seq reads, DNase I hypersensitivity (HS) signal as the density of mapped sequencing tags, and ChIP-seq signal as the density of processed signal enrichment. Tracks 1 to 4 show in black the strand-specific RNA-seq signal in the plus strand or minus strand (denoted to the left of the tracks) of poly(A)− or poly(A)+ RNA from fetal liver TER119+ erythroblasts (ERY). Track 5 depicts in red the signal for DNase I HS, associated with open chromatin, in ERY. Tracks 6 to 8 show in red the ChIP-seq signal for GATA1, TAL1, and KLF1, respectively, in ERY. Tracks 9 and 10 depict the ChIP-seq signal for H3K4me1, enriched along promoter and enhancer regions, in ERY (dark red), and H3K27Ac, associated with active promoters and enhancers, in fetal liver cells (yellow). Peaks of signal enrichment are shown in gray under the DNase I HS track (determined by I-max, empirical FDR <1%) and under the GATA1, TAL1, KLF1, H3K4me1, and H3K27Ac tracks (determined by MACS, empirical FDR <5%). The bottom tracks depict lncRNA transcript models inferred by de novo assembly using Cufflinks (black) and Ensembl gene annotations (red). Shown at the top are the target sites of 3 shRNAs designed against the transcripts from the enhancer site. (B) The alncRNA-EC7 locus is conserved in human. Images from the UCSC Genome Browser depict RNA-seq signal, DNase I HS signal, and ChIP-seq signal in K562 cells as in A. Track 1 shows in blue the non–strand-specific RNA-seq signal of poly(A)+ RNA. Tracks 2 and 3 depict in light blue the ChIP-seq signal for H3K4me1, enriched along promoter and enhancer regions, and H3K27Ac, associated with active promoters and enhancers. Track 4 depicts in dark blue the signal for DNase I HS, associated with open chromatin. Tracks 5 to 8 show in dark blue the ChIP-seq signal for RNA Pol II, GATA1, TAL1, and p300 (enriched at promoter and enhancer sites), respectively. Shown at the bottom are lncRNA transcript models based on our detection of orthologous genomic regions from local alignment and synteny to the mouse genome (black; supplemental Methods) and Ensembl gene annotations (red). The last track depicts in red chromatin interactions associated with binding of the CTCF binding factor, determined by ChIA-PET. (C) Relative abundance of alncRNA-EC7 and Band 3 mRNA across 30 mouse primary tissue and cell types from ENCODE, determined as in Figure 2. Color intensity represents the fractional expression level across all tissues examined. ERY_1 and ERY_2 (red) are TER119+ fetal liver erythroblast biological replicate experiments. (D) (Top) Band 3 expression is coordinated with that of neighboring alncRNA-EC7 during differentiation. (Bottom) Inhibiting alncRNA-EC7 with the shRNAs shown in A abolishes expression of BAND 3 relative to scramble shRNA control (data are mean ± SEM; n = 3).
Discussion
Red blood cell development involves a hierarchy of well-defined cell differentiation states, making it an ideal system to identify lineage- and stage-specific regulators. Ours is the first study to catalog the repertoire of lncRNAs active during erythropoiesis, including >100 previously unannotated lncRNA genes that are often erythroid restricted. We comprehensively characterized these RNAs by their tissue specificity, expression patterns, chromatin state, and TF binding in vivo and integrated these features to select candidates for functional studies. Remarkably, every lncRNA selected this way proved critical for the proper maturation of erythroblasts into specialized erythrocytes. Thus, our study provides a roadmap for the efficient identification of lncRNAs with roles in erythropoiesis, which can be implemented through a useful online resource (http://lodishlab.wi.mit.edu/data/lncRNAs/) where users can select lncRNAs based on their expression and regulation features to discover functional ones.
Recent work has recognized greater cell type specificity in lncRNAs vs mRNAs.8,30 Indeed, many lncRNAs expressed in the mouse fetal liver exhibit strong specificity for 1 or 2 of the 30 cell types examined. Surprisingly, we also find high developmental stage specificity of lncRNAs. We highlight lincRNAs EC-2, -4, and -9, which we find are essentially required for erythrocyte maturation in the fetal liver but are absent in the adult bone marrow. Strikingly, the tissue specificity of certain lncRNAs appears to be lost between the fetal and adult stages of development. Thus, we find crucial differences in the lncRNA programs deployed for the same cell lineage at different stages of development. One explanation for the developmental stage-specific deployment of lncRNAs may be their capacity to mount robust yet short-lived responses to dynamic developmental and environmental cues.
As predicted by their regulated expression and tissue specificity, diverse types of lncRNAs play critical roles during erythropoiesis. Depletion of 5 lincRNAs, 4 alncRNAs, 2 elncRNAs, and 1 shlncRNA severely impaired erythrocyte maturation. Surprisingly, none abolished expression of TER119, an early differentiation marker, indicating that these RNAs are needed during late stages of maturation, when highly specialized processes such as cell size reduction, chromatin condensation, and enucleation take place. Only 3 of these 12 lncRNAs influenced expression of a neighboring gene, akin to similar findings on embryonic stem cell lincRNAs.55 We show that alncRNA-EC7, an enhancer-derived lncRNA, exhibits punctate nuclear localization, is required for activation of neighboring BAND 3, and comes into physical contact with its promoter by chromatin looping in a human cell line, supporting a cis-mediated mechanism. The only shlncRNA examined, EC6, comprises the minimal deleted region in >50% of chronic lymphocytic leukemias56,57 and encodes DLEU2, host to microRNAs 15a and 16-1. A function for DLEU2 independent of microRNA generation had been suggested by the fact that its knockout or ectopic expression shows a stronger phenotype compared with miR-15a/16-1 knockout or misexpression.58-60 Our results now reveal that a predominantly poly(A)− DLEU2 variant is induced during erythropoiesis and promotes red cell maturation at least in part by silencing neighboring genes.
In conclusion, our work demonstrates that lncRNAs of various genomic origins can regulate erythrocyte output, thus contributing to a deeper understanding of the molecular networks driving erythropoiesis that become mutated in disease. Our finding that an enhancer lncRNA is needed for activation of BAND 3, mutated in hereditary hemolytic anemias, predicts a novel disease-relevant locus. Similarly, our data suggest that a variant from the DLEU2 locus, frequently dysregulated in B-cell malignancies, may be an important player during erythropoiesis. These insights highlight lncRNAs as potential therapeutic targets that may be potentially exploited for efficient in vitro production of mature red blood cells.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Laurie Boyer, David Bartel, Nikolai Slavov, Lei Sun, and members of the Lodish laboratory, especially Marko Knoll, for helpful discussions and critical comments on this manuscript. The authors also thank the Massachusetts Institute of Technology and Whitehead Institute flow cytometry cores and the Whitehead Institute genome technology core for technical support and Alec Garcia-Galindo for design and implementation of the online resource accompanying this study.
W.H. is a Merck Fellow of the Life Sciences Research Foundation and is supported by a Pathway to Independence Award (1K99HL118157-01) from the National Institutes of Health, National Heart, Lung, and Blood Institute. This research was supported by the National Institutes of Health, National Cancer Institute Physical Sciences Oncology Center at the Massachusetts Institute of Technology (U54CA143874) and by the grants from National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases DK068348 and from the National Institutes of Health, National Heart, Lung, and Blood Institute, 5P01 HL066105.
Authorship
Contribution: J.R.A.-D., W.H., and H.F.L. designed the research; J.R.A.-D., W.H., J.S., and A.A.G. performed experiments; J.R.A.-D., B.Y., and S.S.P. performed bioinformatics analysis; J.R.A.-D., W.H., B.Y., and J.S. analyzed and interpreted results; J.R.A.-D. and W.H. wrote the manuscript and made the figures; and A.v.O. and H.F.L. provided experimental and analytical insight and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wenqian Hu, Whitehead Institute for Biomedical Research, 9 Cambridge Center, Cambridge, MA 02142; e-mail: whu@wi.mit.edu; and Harvey F. Lodish, Whitehead Institute for Biomedical Research, 9 Cambridge Center, Cambridge, MA 02142; lodish@wi.mit.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal