In this issue of Blood, Stilgenbauer and colleagues report on the prognostic and predictive value of gene mutations assessed prospectively in patients with chronic lymphocytic leukemia (CLL) treated with first-line chemoimmunotherapy.1
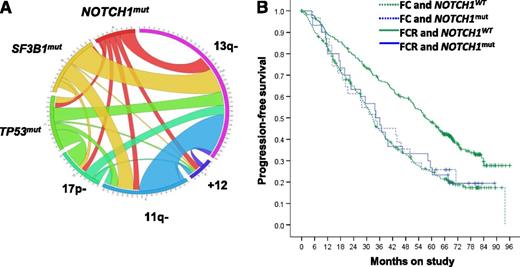
Interrelationship between NOTCH1 mutations (mut) and other recurrently mutated genes and fluorescence in situ hybridization abnormalities in CLL (A). The length of the arc corresponds to the frequency of the genetic lesion, whereas the width of the ribbon indicates the frequency of cooccurrence with the second marker. Kaplan-Meier estimates of PFS for patients stratified by treatment arm and NOTCH1 mutation status (B). WT, wild-type. Panels taken and slightly modified from Figures 1B and 2A in the article by Stilgenbauer et al that begins on page 3247.
Interrelationship between NOTCH1 mutations (mut) and other recurrently mutated genes and fluorescence in situ hybridization abnormalities in CLL (A). The length of the arc corresponds to the frequency of the genetic lesion, whereas the width of the ribbon indicates the frequency of cooccurrence with the second marker. Kaplan-Meier estimates of PFS for patients stratified by treatment arm and NOTCH1 mutation status (B). WT, wild-type. Panels taken and slightly modified from Figures 1B and 2A in the article by Stilgenbauer et al that begins on page 3247.
Heterogeneity in the clinical course of the disease is one of the hallmarks of CLL. The median survival can be <3 years for patients in high-risk subgroups and >25 years in low-risk patients. Understanding the biologic basis for this clinical variability and the development of prognostic markers to dissect the heterogeneity have been areas of intense investigation over the past decades.2 Established prognostic biomarkers include the mutation status of the expressed immunoglobulin gene variable region (IGHV), chromosomal abnormalities assessed by fluorescence in situ hybridization, and expression of CD38, CD49d, and ZAP-70.2 These markers can predict the pace of disease progression in addition to overall survival. As newer and more effective treatment options emerge,3 there is a pressing need to identify biomarkers that can predict how individual patients respond to a specific treatment. In this issue of Blood, Stilgenbauer et al present their analysis on the interaction of somatic mutations and clinical outcomes in a large cohort of prospectively studied patients treated with standard therapy.1 This study may set an example how to obtain the data necessary to tailor treatment to distinct subgroups of patients with CLL.
The CLL 8 trial of the German CLL study group randomized patients to either fludarabine plus cyclophosphamide (FC) or FC with rituximab (FCR) and established the superiority of chemoimmunotherapy.4 Remarkably, of the 817 patients enrolled into the treatment trial, more than 600 could be included in the biomarker analysis with a median follow-up of 70 months. Several acquired somatic mutations have been identified in CLL using next-generation sequencing techniques.5-7 The most common, found in 5% to 15% of patients, affect SF3B1, TP53, and NOTCH1. In the CLL 8 study cohort studied here, at least 1 mutation was identified in 35% of patients, affecting SF3B1 in 18.4%, TP53 in 11.5%, and NOTCH1 in 10%.1 Mutations in TP53 and SF3B1, unmutated IGHV (which identifies the more progressive subtype of CLL), 11q deletion, and 17p deletion were associated with shorter progression-free survival (PFS), whereas TP53 mutations, unmutated IGHV, and 17p deletion were associated with inferior overall survival, consistent with previous observations.
Interestingly, in patients carrying mutated NOTCH1, there was no benefit from the addition of rituximab to FC (see figure).1 Whereas the rate of minimal residual disease–negative remissions in most subgroups was twice as high in FCR-treated patients compared with FC-treated patients, there was no difference in patients with NOTCH1 mutations (50% vs 46.2%). Further, patients with NOTCH1 mutations were the only subgroup that did not demonstrate an improvement in PFS from the addition of rituximab—albeit the difference for patients with mutated TP53 was minimal (median PFS 12.1 months for FC and 15.4 months for FCR).
NOTCH1 was among the first genes identified as recurrently mutated in CLL.5-7 NOTCH1 is a ligand-activated transcription factor that regulates downstream pathways important for cellular growth and plays a key role in T-cell acute lymphoblastic leukemia. Most of the mutations found in CLL are frameshift mutations that lead to a truncated constitutively active protein. Although the role of activated NOTCH1 in the pathobiology of CLL remains to be defined, more rapid disease progression and inferior survival in patients with NOTCH1 mutations have been reported.5,6,8,9 Consistent with a postulated role in driving disease progression is the increasing prevalence of NOTCH1 mutations in chemotherapy-refractory patients and in patients with Richter transformation.5,6
As the observation that mutations in NOTCH1 may predict a lack of benefit from rituximab awaits confirmation, it will also be important to investigate whether mutated NOTCH1 affects the treatment outcome with other anti-CD20 antibodies or monoclonal antibodies in general. Uncovering the mechanism of how NOTCH1 mutations influence response to rituximab will also require further study; in the CLL 8 trial there was no association with lower CD20 expression, more advanced disease or absolute lymphocyte count.1 If confirmed, this raises the intriguing possibility that a better understanding of the molecular pathways downstream of NOTCH1 could uncover novel mechanisms of resistance to antibody therapy. From a therapeutic standpoint, patients with NOTCH1 mutations might benefit from tailored approaches including agents that inhibit NOTCH1 activation or kinase inhibitors that target B-cell receptor signaling. The latter is suggested by the observation that NOTCH1 mutations, trisomy 12, and a specific B-cell receptor configuration (referred to as subset 8) appear to cooperate in Richter transformation.10
In summary, 17p deletion and TP53 mutations predicted a particularly poor outcome with chemoimmunotherapy, mutated NOTCH1 was associated with no benefit from the addition of rituximab to chemotherapy, and SF3B1 mutations, although neutral in regard to treatment response, were associated with more rapid disease progression in this prospective cohort of patients treated according to standard criteria. Whether newer treatments can overcome the negative impact of these mutations remains to be determined, but emerging data with novel agents are promising,3 and enrollment of patients into clinical trials that aim to address these fundamental translational questions will be critical.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal