Key Points
ATF3 inhibits lipopolysaccharide-driven CXCL1 production by airway epithelia.
ATF3 controls neutrophil recruitment to the wild-type lung and chemotaxis in vitro via TIAM2 expression.
Abstract
Expression of the activating transcription factor 3 (ATF3) gene is induced by Toll-like receptor (TLR) signaling. In turn, ATF3 protein inhibits the expression of various TLR-driven proinflammatory genes. Given its counter-regulatory role in diverse innate immune responses, we defined the effects of ATF3 on neutrophilic airway inflammation in mice. ATF3 deletion was associated with increased lipopolysaccharide (LPS)-driven airway epithelia production of CXCL1, but not CXCL2, findings concordant with a consensus ATF3-binding site identified solely in the Cxcl1 promoter. Unexpectedly, ATF3-deficient mice did not exhibit increased airway neutrophilia after LPS challenge. Bone marrow chimeras revealed a specific reduction in ATF3−/− neutrophil recruitment to wild-type lungs. In vitro, ATF3−/− neutrophils exhibited a profound chemotaxis defect. Global gene expression analysis identified ablated Tiam2 expression in ATF3−/− neutrophils. TIAM2 regulates cellular motility by activating Rac1-mediated focal adhesion disassembly. Notably, ATF3−/− and ATF3-sufficient TIAM2 knockdown neutrophils, both lacking TIAM2, exhibited increased focal complex area, along with excessive CD11b-mediated F-actin polymerization. Together, our data describe a dichotomous role for ATF3-mediated regulation of neutrophilic responses: inhibition of neutrophil chemokine production but promotion of neutrophil chemotaxis.
Introduction
Activating transcription factor 3 (ATF3) is a counter-regulatory immune transcription factor (TF). It is induced by Toll-like receptor (TLR) signaling and, in turn, inhibits the transcription of genes encoding diverse TLR-driven, proinflammatory mediators.1,2 ATF3 is a member of the ATF/CREB family of basic leucine zipper TFs.3 ATF3 homodimers inhibit gene targets directly through association with histone deacetylase 1,1,4 whereas ATF3-containing heterodimers positively regulate gene expression.5-8 Basal ATF3 expression is low in most cell types, yet is rapidly and transiently elevated in response to various stimuli,9 in part due to an autoregulatory ATF3 binding site in its promoter.10
Transient TLR-driven signals induce nuclear factor κB (NF-κB)–mediated ATF3 transcription, which then attenuates the expression of diverse other NF-κB targets.4 Persistent TLR stimulation, however, additionally transcribes the NF-κB enhancer, C/EBP-δ, which augments NF-κB signaling and overcomes ATF3-mediated inhibition.4 ATF3 induction and function on immune responses are highly cell type and stimulation dependent.9,11 ATF3 is necessary to restrain immune activation and immunopathology in models of sepsis1 and lipopolysaccharide (LPS)-induced febrile responses12 ; however, attenuation of NK effector responses by ATF3 contributes to increased murine cytomegalovirus-mediated liver damage.11 Therefore, understanding the immunoregulatory role(s) of ATF3 demands careful evaluation in specific immune contexts.
Lung challenge with diverse microbial products induces production of glutamate-leucine-arginine (ELR+) chemokine family members, including CXCL1, by resident parenchymal13-16 and hematopoietic cells.16-18 ELR+ chemokines are highly neutrophil specific. Their transcription, production, tissue localization,19 and neutralization20 are tightly regulated. Circulating neutrophils sample chemokines close to the endothelial wall while rolling on activated endothelia. Chemokine receptor ligation induces conformational changes in neutrophil β2 integrins, allowing firm adhesion to the endothelia and eventual extravasation. Extravasation and chemotaxis require the coordinated assembly and disassembly of cytoskeletal and adhesive structures. In neutrophils, multiprotein focal complexes link the intracellular cytoskeleton to integrin-mediated extracellular structures. These highly dynamic structures are required for proper cell motility; however, they are incompletely understood.21
Herein, we describe a novel and complex role for ATF3 regulation of neutrophil migration. LPS challenge resulted in significantly increased production of the potent neutrophil chemoattractant, CXCL1, in ATF3−/− mice; however, neutrophil recruitment was not increased in ATF3−/− lungs. In vitro studies revealed significantly impaired ATF3−/− neutrophil migration to chemokine gradients. Gene expression analysis revealed ablation of Tiam2, a focal adhesion regulator, in mature ATF3−/− neutrophils. This was associated with dysregulated adhesion structures and cytoskeletal organization in ATF3−/−, compared with wild-type (WT), neutrophils. Further, TIAM2 knockdown in WT neutrophils phenocopied the ATF3−/− neutrophil chemotaxis defect. Together, these studies describe a complex and previously unappreciated role for ATF3 regulation of neutrophil recruitment to the lung and neutrophil chemotaxis generally.
Materials and methods
Mice and in vivo studies
ATF3−/− mice,22 backcrossed >10 generations to a C57Bl/6NJ background, and WT C57Bl/6NJ mice (Taconic Farms) were maintained in the Cincinnati Children’s Hospital Medical Center’s specific pathogen-free animal facility. Intratracheal (i.t.) challenge studies were performed as described23 using phosphate-buffered saline (Gibco), ultrapure LPS (Invivogen), recombinant (r)CXCL1 (PeproTech), and/or anti-CXCL1 or isotype control antibodies (R&D Systems). For bone marrow (BM) transplants, WT CD45.1+ mice (B6.SJL-Ptprca/BoyAiTac; Taconic Farms) were lethally irradiated (700 and 475 rad, separated by 4 hours), and rescued with 2 × 106 BM cells from CD45.2+ ATF3−/− or WT mice. Bronchoalveolar lavage (BAL) was performed with 500 μL Hanks balanced salt solution (Gibco), and lungs were harvested into TRIzol (Invitrogen). BAL fluid (BALF) cytospins were analyzed by Diff-Quik staining, and cell-free supernatant was assayed for CXCL1, CXCL2, or CXCL5 (R&D Systems) by enzyme-linked immunosorbent assay (ELISA). Experimental procedures were approved by the Cincinnati Children's Hospital Medical Center's Institutional Animal Care and Use Committees.
In vitro mouse tracheal epithelial cell culture
Mouse tracheal epithelial cells (MTECs) were isolated and cultured as described.24 Briefly, tracheas from 4- to 5-week-old mice were harvested and disaggregated by 0.1% pronase (Roche) and DNase I (Sigma-Aldrich) digestion, followed by fibroblast removal by plastic adherence. MTECs were then cultured on type I collagen-coated, 0.4-μm pore transwell inserts (BD Biosciences) until formation of tight junctions (R > 1000 Ω; 10-14 days). MTECs were stimulated apically with LPS. Basolateral cytokine production was quantified by ELISA.
mRNA analysis
Neutrophils were isolated as described,25 and RNA was harvested by TRIzol. Atf3, Cxcl1, Cxcl2, Tiam1, and Tiam2 mRNA, normalized to β-actin expression, were quantified by quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) (LightCycler 480; Roche), using the following primers: β-actin, 5′-GGCCCAGAGCAAGAGAGGTA-3′, 5′-GGTTGGCCTTAGGGTTCAGG-3′; Atf3, 5′-AGCCTGGAGCAAAATGATGCTT-3′, 5′-AGGTTAGCAAAATCCTCAAACAC-3′; Cxcl1, 5′-ACCCAAACCGAAGTCATAGC-3′, 5′-TCTCCGTTACTTGGGGACAC-3′; Cxcl2, 5′-TCCAGGTCAGTTAGCCTTGC-3′, 5′-CGGTCAAAAAGTTTGCCTTG-3′; Tiam1, 5′-TCACTCAGGACTTGAGCAGC-3′, 5′-TGGGAGAATGTGCCAGAAAC-3′; and Tiam2, 5′-CAGGGAAAAGATGGAGCAGA-3′, 5′-ATGGCTCTCTGTTGGTGCTT-3′. For microarray analysis, WT and ATF3−/− neutrophils were isolated by Ly6G+ immunomagnetic selection (Miltenyi), and RNA was harvested by RNeasy Mini Kit (Qiagen). RNA was analyzed on an Agilent SurePrint G3 Mouse GE 8x60K microarray with an Agilent 2100 Bioanalyzer (data accession no. GSE53973).
Immunofluorescence and flow cytometry
Adherent neutrophils (on uncoated or fibrinogen-coated [25 μg/mL; Sigma-Aldrich] slides) or neutrophils suspended in Hanks balanced salt solution + 0.1% bovine serum albumin (Sigma-Aldrich) were stimulated with 10 μM Formyl-Methionyl-Leucyl-Phenylalanine (fMLP) (Sigma-Aldrich), 1 mM Ca2+/Mg2+ for 10 minutes, and fixed with 2% to 4% paraformaldehyde for 20 minutes. Cells were permeabilized with 0.1% Triton X-100 or 0.03% saponin (ACROS Organics) and stained with rhodamine-phalloidin (1:40; Invitrogen) and/or α-vinculin (1:50; Sigma-Aldrich). Adherent cells were mounted in SlowFade Gold (Invitrogen) and imaged on a Leica DMI6000 fluorescence microscope using a 63×/1.3 NA objective, with an ORCA-ERC4742-95 camera (Hamamatsu) driven by Openlab (version 5.5.0) software (Perkin Elmer). Flow cytometric analysis of nonadherent neutrophils was performed by incubation with monoclonal antibodies to CD11a, CD11b, and CD18 (1:100; eBioscience), followed by use of an LSR II flow cytometer (BD Biosciences).
In vitro neutrophil migration assays
Time-lapse video microscopy was performed as described previously.25 Using a Zigmond chamber (NeuroProbe), adherent neutrophils were imaged every 5 seconds using ImageJ 1.43J software on a Zeiss Axiovert 200 microscope at 10×/0.3 NA objective, equipped with an ORCA-ER-C4742-95 camera (Hamamatsu) driven by Openlab software (version 5.5.0), in a gradient of 10 μM fMLP in Hanks balanced salt solution + 10 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (Gibco) for 20 minutes in a 37°C chamber. Individual neutrophil paths were traced in ImageJ software, version 1.45s.
Lentivirus-mediated shRNA knockdown of Tiam2
Hematopoietic stem cells from WT mice were transduced with lentiviral constructs (Sigma-Aldrich) expressing short hairpin RNA (shRNA) against Tiam2 or a scrambled control. Neutrophils were generated in culture and used for experiments as previously described.26
Statistics
Prism GraphPad 5 software was used for statistical analysis using unpaired 2-tailed Student t tests or 1-way or 2-way ANOVA tests as appropriate. Microarray gene targets were analyzed using 2 independent approaches: (1) a 10% false discovery rate27 cutoff was used for nonparametric significance analysis of microarrays28 performed in R with the Bioconductor Siggenes package, and (2) probe expression was normalized by robust microarray average in GeneSpring (Agilent), and probes with absolute signal fold change >2 were evaluated by an unpaired Student t test and corrected for multiple comparisons by the Benjamini-Hochberg post test.
Results
ATF3 regulates LPS-driven CXCL1 production in the airway
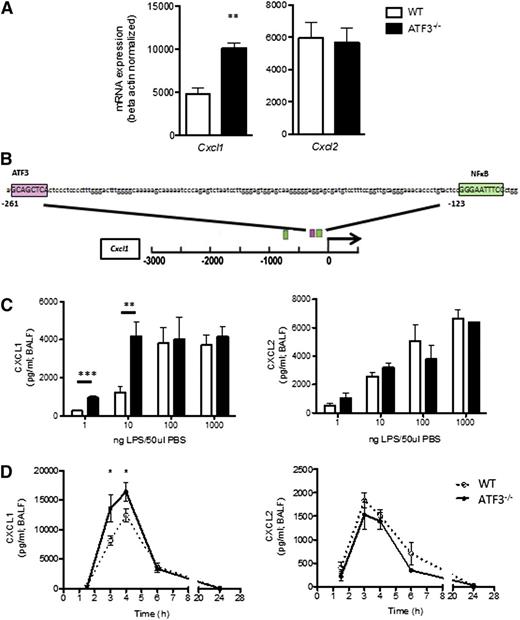
To evaluate the role of ATF3 in acute lung inflammation, we analyzed lung homogenates for ELR+ chemokine transcripts. Airway LPS challenge induced expression of both Cxcl1 and Cxcl2, yet only Cxcl1 transcripts were significantly increased in ATF3−/− lungs compared with WT lungs (Figure 1A). Cxcl1 promoter analysis revealed a consensus ATF3 binding site (Figure 1B) absent from the Cxcl2 promoter (data not shown), suggesting specific ATF3 regulation of Cxcl1 expression.
ATF3 regulates LPS-induced airway CXCL1 production. (A) WT (white bars) and ATF3−/− (black bars) mice were challenged i.t. with 10 ng LPS and lavaged 4 hours later. Following lavage, the left upper lung lobes were removed for mRNA quantification by qRT-PCR for (left) Cxcl1 or (right) Cxcl2. Representative of a single experiment, N = 3 to 5 mice/group. (B) The −3-kb region of the Cxcl1 promoter was analyzed using online TESS software to identify the putative ATF3 binding sequence GCA CGT CA (pink box), as well as known NF-κB binding sites (green boxes). (C) WT (white bars) and ATF3−/− (black bars) mice were challenged i.t. with increasing doses of LPS. After 2 hours, lungs were lavaged, and cell-free supernatants were analyzed by ELISA for (left) CXCL1 or (right) CXCL2. Representative of 3 experiments, N = 2 to 9 mice/group. (D) WT (dotted lines) and ATF3−/− (solid lines) mice challenged i.t. with 10 ng LPS were lavaged at the indicated times to evaluate (left) CXCL1 or (right) CXCL2 production by ELISA. Representative of 7 experiments; N = 3 to 7 mice/group. Statistics are unpaired 2-tailed Student t test compared with WT control. Data are mean ± standard error of the mean (SEM). *P < .05, **P < .01.
ATF3 regulates LPS-induced airway CXCL1 production. (A) WT (white bars) and ATF3−/− (black bars) mice were challenged i.t. with 10 ng LPS and lavaged 4 hours later. Following lavage, the left upper lung lobes were removed for mRNA quantification by qRT-PCR for (left) Cxcl1 or (right) Cxcl2. Representative of a single experiment, N = 3 to 5 mice/group. (B) The −3-kb region of the Cxcl1 promoter was analyzed using online TESS software to identify the putative ATF3 binding sequence GCA CGT CA (pink box), as well as known NF-κB binding sites (green boxes). (C) WT (white bars) and ATF3−/− (black bars) mice were challenged i.t. with increasing doses of LPS. After 2 hours, lungs were lavaged, and cell-free supernatants were analyzed by ELISA for (left) CXCL1 or (right) CXCL2. Representative of 3 experiments, N = 2 to 9 mice/group. (D) WT (dotted lines) and ATF3−/− (solid lines) mice challenged i.t. with 10 ng LPS were lavaged at the indicated times to evaluate (left) CXCL1 or (right) CXCL2 production by ELISA. Representative of 7 experiments; N = 3 to 7 mice/group. Statistics are unpaired 2-tailed Student t test compared with WT control. Data are mean ± standard error of the mean (SEM). *P < .05, **P < .01.
ATF3 functions are complex and context dependent. ATF3 inhibits immune responses to transient signals; however, following persistent TLR signaling, ATF3 silences its expression via autoinhibition.4 ATF3 regulation is also stimulus dose dependent, as in LPS stimulation of ATF3−/− dendritic cells.2 We therefore challenged WT and ATF3−/− mice with increasing LPS doses. Peripheral blood counts were similar between the groups before and after challenge (data not shown). We found that ATF3 deficiency resulted in significantly increased airway CXCL1 production at low LPS doses (1 and 10 ng/mL), an effect that was overcome at higher doses of LPS. LPS-induced ATF3 regulation was CXCL1 specific, as CXCL2 production did not vary by genotype at any LPS dose (Figure 1C). No genotype-specific differences were observed in the (very low) concentrations of CXCL1 and CXCL2 measurable following control phosphate-buffered saline challenge (data not shown). Kinetic analysis of low-dose LPS challenge revealed that induction of CXCL1 and CXCL2 production in WT airways was rapid and transient, returning to baseline levels within 24 hours. ATF3−/− mice had a similar temporal profile of CXCL1 production, although concentrations were significantly elevated at 3 (twofold) and 4 hours (1.3-fold) compared with WT controls (Figure 1D). Again, the ATF3 effect was CXCL1 specific, as there were no significant differences in the production of CXCL2 (Figure 1D) or CXCL5 (supplemental Figure 1 on the Blood Web site). Together, these data suggest a previously unappreciated role for ATF3 in the regulation of LPS-induced CXCL1 production during acute lung inflammation.
Resident lung cells are the source of increased CXCL1 production in ATF3−/− mice
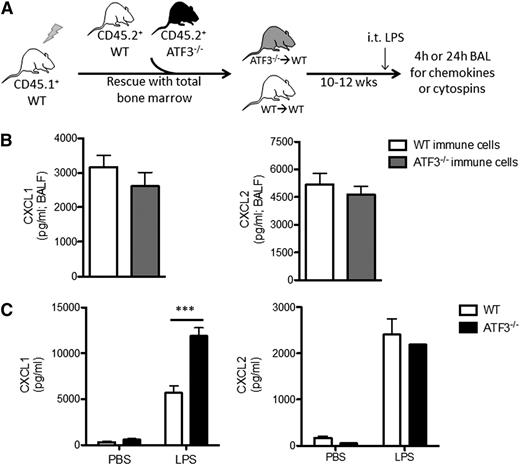
LPS stimulation induces CXCL1 production by many cell types in the lung, including endothelial cells,13 type 2 epithelial and Clara cells,14-16,29 and various resident and recruited BM-derived cells, such as alveolar macrophages16,17,29 and neutrophils.18,30 To test whether recruited BM-derived cells are the major source for differential CXCL1 production, we adoptively transplanted BM cells from CD45.2+ WT or ATF3−/− mice into CD45.1+ WT recipients (Figure 2A). Reconstitution efficiencies and peripheral blood cell populations were equivalent between transplanted groups prior to LPS challenge (supplemental Figure 2). LPS challenge (i.t.) induced similar CXCL1 and CXCL2 production in both groups (Figure 2B), indicating that recruited BM-derived cells are likely not the source of increased airway CXCL1 in ATF3−/− mice.
Lung parenchymal cells, not recruited hematopoietic cells, are essential to ATF3 regulation of chemokine production. (A) BM transplantation schema. CD45.1+ WT mice were lethally irradiated and reconstituted with BM from CD45.2+ WT (white) or ATF3−/− (black) mice. Ten to 12 weeks after transplant, CD45.1+ WT mice with CD45.2+ WT (white) or ATF3−/− (gray) BM were used for experimentation. (B) LPS challenge (i.t.) and BAL fluid determination of (left) CXCL1 or (right) CXCL2 by ELISA at 4 hours in CD45.1+ WT mice reconstituted with CD45.2+ congenic WT (white bars) or ATF3−/− (black bars) BM cells. Pooled from 3 independent experiments; N = 19 to 20 ATF3−/− donor conditions and 14 to 15 WT donor conditions. Statistics are unpaired 2-tailed Student t test. (C) MTECs from WT (white bars) or ATF3−/− (black bars) mice were challenged apically with 10 ng/mL LPS for 18 hours. Basolateral media were then analyzed for (left) CXCL1 or (right) CXCL2 production by ELISA. Representative of 3 experiments; N = 2 to 3 wells/condition. Statistics are 2-way ANOVA with Bonferroni correction. Data are mean ± SEM. ***P < .001.
Lung parenchymal cells, not recruited hematopoietic cells, are essential to ATF3 regulation of chemokine production. (A) BM transplantation schema. CD45.1+ WT mice were lethally irradiated and reconstituted with BM from CD45.2+ WT (white) or ATF3−/− (black) mice. Ten to 12 weeks after transplant, CD45.1+ WT mice with CD45.2+ WT (white) or ATF3−/− (gray) BM were used for experimentation. (B) LPS challenge (i.t.) and BAL fluid determination of (left) CXCL1 or (right) CXCL2 by ELISA at 4 hours in CD45.1+ WT mice reconstituted with CD45.2+ congenic WT (white bars) or ATF3−/− (black bars) BM cells. Pooled from 3 independent experiments; N = 19 to 20 ATF3−/− donor conditions and 14 to 15 WT donor conditions. Statistics are unpaired 2-tailed Student t test. (C) MTECs from WT (white bars) or ATF3−/− (black bars) mice were challenged apically with 10 ng/mL LPS for 18 hours. Basolateral media were then analyzed for (left) CXCL1 or (right) CXCL2 production by ELISA. Representative of 3 experiments; N = 2 to 3 wells/condition. Statistics are 2-way ANOVA with Bonferroni correction. Data are mean ± SEM. ***P < .001.
Epithelial cells are among the first and most abundant cells exposed to microbial products in the lung and play a critical role in initiating immune responses.14-16,29 To determine the contribution of lung epithelium to ATF3-mediated CXCL1 regulation, we stimulated MTECs from WT and ATF3−/− mice with LPS. Apical LPS stimulation resulted in a significant increase in basolateral CXCL1 secretion in ATF3−/− compared with WT and MTEC cultures. This regulatory ATF3 effect was again restricted to CXCL1, as CXCL2 secretion was equivalent between genotypes (Figure 2C). Together, these results suggest that resident lung epithelial cells, not recruited or resident BM-derived cells, are the likely locus of ATF3-mediated regulation of LPS-driven CXCL1 in the airway.
ATF3−/− neutrophils exhibit impaired lung recruitment
CXCL1 induces neutrophil chemotaxis, but CXCL2 and CXCL5, also induced by LPS challenge of the airway, are additional neutrophil chemoattractants. Therefore, we sought to better define the specific role of CXCL1 in LPS-driven neutrophil lung recruitment: i.t. administration of rCXCL1 was sufficient to recruit WT neutrophils to the lung in a dose-dependent manner (Figure 3A). Coadministration of CXCL1-neutralizing antibodies with LPS significantly reduced bioavailable CXCL1 without affecting LPS-induced Cxcl1 transcription or CXCL2 production (supplemental Figure 3). Remarkably, CXCL1 neutralization attenuated LPS-induced neutrophil recruitment by almost 75% (Figure 3B), indicating that CXCL1 plays a dominant role in neutrophil recruitment in response to low-dose LPS challenge of the airway.
ATF3−/−neutrophil lung recruitment in impaired. (A) BAL fluid from WT mice challenged with increasing amounts of rCXCL1 i.t. were evaluated to determine neutrophil recruitment to the lung after 18 to 24 hours by microscopic evaluation of cytospins. Representative of 2 independent experiments, N = 3 to 5 mice/group. Statistics are 1-way ANOVA. (B) LPS (10 ng) and 50 μg isotype control (white bars) or neutralizing CXCL1 antibody (black bars), were coadministered i.t. to WT mice. Neutrophil content of BALF was determined as in A. Representative of 2 independent experiments; N = 3 to 5 mice/group. Statistics are unpaired 2-tailed Student t test. (C) WT (dotted lines) and ATF3−/− (black lines) mice were challenged i.t. with 10 ng LPS and neutrophils enumerated as in A. Representative of 8 independent experiments; N = 3 to 7 mice/group. (D) BM chimeras from Figure 2A were challenged and neutrophil recruitment determined as in A. Representative of 3 independent experiments; N = 14 to 16 mice/group. Statistics are unpaired Student 2-tailed t test. Data are mean ± SEM. *P < .05, ***P < .001.
ATF3−/−neutrophil lung recruitment in impaired. (A) BAL fluid from WT mice challenged with increasing amounts of rCXCL1 i.t. were evaluated to determine neutrophil recruitment to the lung after 18 to 24 hours by microscopic evaluation of cytospins. Representative of 2 independent experiments, N = 3 to 5 mice/group. Statistics are 1-way ANOVA. (B) LPS (10 ng) and 50 μg isotype control (white bars) or neutralizing CXCL1 antibody (black bars), were coadministered i.t. to WT mice. Neutrophil content of BALF was determined as in A. Representative of 2 independent experiments; N = 3 to 5 mice/group. Statistics are unpaired 2-tailed Student t test. (C) WT (dotted lines) and ATF3−/− (black lines) mice were challenged i.t. with 10 ng LPS and neutrophils enumerated as in A. Representative of 8 independent experiments; N = 3 to 7 mice/group. (D) BM chimeras from Figure 2A were challenged and neutrophil recruitment determined as in A. Representative of 3 independent experiments; N = 14 to 16 mice/group. Statistics are unpaired Student 2-tailed t test. Data are mean ± SEM. *P < .05, ***P < .001.
Given the elevated CXCL1 levels observed after LPS challenge of ATF3−/− airways and the importance of CXCL1 in this model, we expected increased LPS-driven neutrophil recruitment to ATF3−/− airways. Unexpectedly, we observed no differences between ATF3−/− and WT airway neutrophil numbers during the 24 hours after i.t. LPS challenge (Figure 3C). Additionally, there were no observed differences in the number of recruited monocyte/macrophages, lymphocytes, eosinophils, or denuded epithelial cells (data not shown). This could not be explained by compensatory increases in other ELR+ chemokines (Figure 1C-D; supplemental Figure 1), or reduced numbers or altered tissue distribution of ATF3−/− neutrophils (supplemental Figure 4). We thus examined potential neutrophil-intrinsic effects of ATF3 on neutrophil recruitment, using WT recipients reconstituted with WT or ATF3−/− BM cells, as in Figure 2A. Interestingly, we observed a striking defect in the recruitment of CD45.2+ ATF3−/− neutrophils to CD45.1+ WT airways; recruitment was reduced by almost 50% compared with WT controls (Figure 3D).
ATF3−/− neutrophils exhibit impaired in vitro migration
As shown in supplemental Figure 5, CXCL1 receptor (CXCR2) expression was comparable between ATF3−/− and WT neutrophils, suggesting (albeit not proving) that ATF3−/− neutrophils can respond to CXCL1. This prompted us to hypothesize that the above in vivo recruitment defect might be due to defects in the migration machinery of ATF3−/− neutrophils. We therefore compared WT and ATF3−/− neutrophil migration in vitro in response to fMLP, a well-established neutrophil chemoattractant, using time-lapse microscopy. WT neutrophils readily assumed a polarized morphology (Figure 4A) and migrated up fMLP gradients (Figure 4B). ATF3−/− neutrophils were also able to polarize and generate lamellipodial protrusions (Figure 4A). Notably, however, ATF3−/− neutrophils exhibited a striking inability to translocate, as significantly fewer ATF3−/− neutrophils moved productively (movement >20 μm) compared with WT neutrophils (88% WT vs 10% ATF3−/−; Figure 4A). Supplemental movies clearly demonstrate this ATF3−/− migration defect, as well as their preserved ability to generate a polarized morphology. Of the few ATF3−/− neutrophils that did translocate, their ability to migrate was severely impaired, with a significant reduction (80%) in the distance traveled compared with WT neutrophils (Figure 4B; supplemental Movies 1 and 2).
ATF3−/−neutrophils exhibit a profound defect in directional motion. (A) Representative images (1 minute between each frame) of migrating (upper) WT or (lower) ATF3−/− neutrophils in a gradient of fMLP on uncoated surfaces in a Zigmond chamber. fMLP concentration increases from left to right. Images were captured every 5 seconds for 20 minutes at 37°C. Original magnification, ×100. The number of neutrophils able to move productively (20 μm from their starting position) is quantified at right. Images are representative of, and graph is pooled from, 4 independent experiments; N = 6 WT and 9 ATF3−/− independent acquisitions/experiment, and 20 to 100 neutrophils were evaluated/acquisition. Images were captured using a Zeiss Axiovert 200 microscope at 10×/0.3 NA objective, equipped with an ORCA-ER C4742-95 camera driven by Openlab (version 5.5.0) software. (B) Paths of migrating (left) WT and (right) ATF3−/− neutrophils were traced using ImageJ software. The schema represent cells moving in fMLP gradient over 20 minutes. The total distance traveled from the origin (net translocation) is quantified at right. Schema are representative of, and graph is pooled from, 3 independent experiments; N = 4 WT and 9 ATF3−/− independent acquisitions/experiment. Statistics are unpaired 2-tailed Student t test. Data are mean ± SEM. **P < .01, ***P < .001.
ATF3−/−neutrophils exhibit a profound defect in directional motion. (A) Representative images (1 minute between each frame) of migrating (upper) WT or (lower) ATF3−/− neutrophils in a gradient of fMLP on uncoated surfaces in a Zigmond chamber. fMLP concentration increases from left to right. Images were captured every 5 seconds for 20 minutes at 37°C. Original magnification, ×100. The number of neutrophils able to move productively (20 μm from their starting position) is quantified at right. Images are representative of, and graph is pooled from, 4 independent experiments; N = 6 WT and 9 ATF3−/− independent acquisitions/experiment, and 20 to 100 neutrophils were evaluated/acquisition. Images were captured using a Zeiss Axiovert 200 microscope at 10×/0.3 NA objective, equipped with an ORCA-ER C4742-95 camera driven by Openlab (version 5.5.0) software. (B) Paths of migrating (left) WT and (right) ATF3−/− neutrophils were traced using ImageJ software. The schema represent cells moving in fMLP gradient over 20 minutes. The total distance traveled from the origin (net translocation) is quantified at right. Schema are representative of, and graph is pooled from, 3 independent experiments; N = 4 WT and 9 ATF3−/− independent acquisitions/experiment. Statistics are unpaired 2-tailed Student t test. Data are mean ± SEM. **P < .01, ***P < .001.
ATF3 induction during neutrophil development is necessary for TIAM2 expression
This cell-intrinsic ATF3 effect in neutrophil function was initially surprising due to 2 observations. First, stimulus-dependent induction of ATF3 mRNA takes ∼1 hour,9 whereas ATF3−/− neutrophil migration defects occur within minutes; therefore, ATF3-dependent chemotaxis effects are likely not due to de novo ATF3 transcription and regulation of gene expression. Second, we detected neither ATF3 protein (data not shown) nor mRNA (supplemental Figure 6) in mature WT neutrophils, either unstimulated or following LPS stimulation, indicating that ATF3 expression is extremely low or absent in mature neutrophils. Thus, the intrinsic migratory defects of ATF3−/− neutrophils are likely a function of events occurring prior to neutrophil maturation, including control of target gene expression during myeloid differentiation.31
We thus sought to determine potential gene targets regulated by ATF3 during neutrophil development by comparing global gene expression in unstimulated WT and ATF3−/− neutrophils. We detected significant differences in only 9 protein-coding genes: Tiam2, Erdr1, Odz3, Igh-VJ558, Igj, Efna2, Synpo, Susd4, and Mtus2. Tiam2 was the most robust and statistically significant differentially expressed gene, with a 284-fold reduction in ATF3−/− neutrophils as quantified by microarray (corrected P < .0001; supplemental Table 1), and a 99% reduction by qRT-PCR (Figure 5A). Tiam2 was also the only microarray-implicated gene whose product is purported to affect cell migration.21 Consistent with potential direct regulation of Tiam2 by ATF3, both human and mouse TIAM2 promoters contain consensus ATF3 binding sites (Figure 5B). TIAM1 is closely related to TIAM2, yet it does not compensate functionally for TIAM2 reduction,21 nor does it contain an ATF3 binding site (data not shown). Additionally, TIAM1 mRNA was not differentially expressed in TIAM2-lacking ATF3−/− neutrophils as quantified by microarray (fold change = −1.003, corrected P = .9889) or by qRT-PCR (Figure 5A).
ATF3 regulates Tiam2 expression during neutrophil development. (A) RNA from WT (white bars) or ATF3−/− (black bars) neutrophils were analyzed by qRT-PCR for Tiam2 or Tiam1 expression. Representative of 3 independent experiments; N = 2 to 6 mice/genotype. Statistics are unpaired 2-tailed Student t test. (B) The −3-kb region of Tiam2 variant 1 (NM_011878) shown with the consensus ATF3 binding site TGA CGC CA (pink box) indicated relative to the known transcriptional activator NF-kB binding site (green box). (C) mRNA expression of Tiam2 during differentiation of WT myeloid progenitors (c-Kit+Lineage− BM cells) to mature neutrophils. Bar graph shows Tiam2 expression at indicated time in culture. (D) Immunoblot of Tiam2 protein expression during WT LK differentiation as in C. (E) Immunoblot of Tiam2 protein expression in WT and ATF3−/− neutrophils.
ATF3 regulates Tiam2 expression during neutrophil development. (A) RNA from WT (white bars) or ATF3−/− (black bars) neutrophils were analyzed by qRT-PCR for Tiam2 or Tiam1 expression. Representative of 3 independent experiments; N = 2 to 6 mice/genotype. Statistics are unpaired 2-tailed Student t test. (B) The −3-kb region of Tiam2 variant 1 (NM_011878) shown with the consensus ATF3 binding site TGA CGC CA (pink box) indicated relative to the known transcriptional activator NF-kB binding site (green box). (C) mRNA expression of Tiam2 during differentiation of WT myeloid progenitors (c-Kit+Lineage− BM cells) to mature neutrophils. Bar graph shows Tiam2 expression at indicated time in culture. (D) Immunoblot of Tiam2 protein expression during WT LK differentiation as in C. (E) Immunoblot of Tiam2 protein expression in WT and ATF3−/− neutrophils.
Further, TIAM2 mRNA and protein expression increased during myeloid cell differentiation to neutrophils (Figure 5C-D), yet TIAM2 protein expression was almost absent in ATF3−/− neutrophils compared with WT (Figure 5E). Together, these studies suggest that ATF3 expression during neutrophil differentiation is required for proper expression of TIAM2 in mature neutrophils.
Adhesion structures and cytoskeletal organization are abnormal in stimulated ATF3−/− neutrophils
Focal complexes link static integrin-mediated extracellular interactions to the dynamic cellular cytoskeleton, and proper assembly and disassembly of adhesive structures are necessary for cellular movement. Given the role of TIAM2 in focal adhesion (FA) disassembly and cellular motility21 and the lack of TIAM2 in ATF3−/− neutrophils, we hypothesized that abnormal adhesive structure disassembly could underlie impaired ATF3−/− neutrophil chemotaxis. Indeed, fMLP-stimulated ATF3−/− neutrophils exhibited abnormally intense focal complex staining compared with WT neutrophils (Figure 6A). Adhesive structures were significantly larger (Figure 6B) and more numerous (Figure 6C) in ATF3−/− neutrophils. Adhesive structures can regulate F-actin polymerization.32 Therefore, we also evaluated stimulation-dependent F-actin polymerization. Compared with WT neutrophils, ATF3−/− neutrophils displayed aberrant polymerized F-actin morphology (Figure 6A). ATF3−/− neutrophils exhibited broader and thicker polymerized F-actin-staining lamellipodia, resulting in significantly increased areas of polymerized F-actin (Figure 6D). Increased ATF3−/− neutrophil F-actin polymerization was adhesion dependent, as there were no differences between ATF3−/− and WT neutrophils stimulated in suspension (supplemental Figure 7). Further, increased ATF3−/− F-actin polymerization was CD11b mediated, as demonstrated using the CD11b-specific ligand fibrinogen (Figure 6D). Expression of the principal neutrophil β2 integrins CD11a, CD11b, and CD18 was normal (supplemental Figure 8). These data suggest that impaired TIAM2-mediated focal complex disassembly in ATF3−/− neutrophils is a potential mechanism of impaired ATF3−/− neutrophil chemotaxis.
ATF3−/−neutrophils lacking TIAM2 exhibit dysregulated integrin-dependent cytoskeletal organization and adhesive structure regulation. (A) (Upper) WT or (lower) ATF3−/− neutrophils were allowed to adhere to uncoated glass slides and then stimulated with 10 μM fMLP and stained for polymerized F-actin by phalloidin-rhodamine and focal complexes (arrows) and focal contacts (arrowheads) by antivinculin-Alexa Fluor 488. Representative focal complex/contacts (green), polymerized F-actin (red), 4′6 diamidino-2-phenylindole–stained nuclei (blue), and merged immunofluorescent images are shown. Original magnification, ×630. Representative of 2 (vinculin) or 5 (phalloidin) independent experiments. Fluorescence images were captured at room temperature using a Leica DMI6000 fluorescence microscope at 63×/1.3 NA objective with an ORCA-ER C4742-95 camera driven by Openlab (version 5.5.0) software. Total (B) area and (C) numbers of vinculin-containing focal structures quantified from A for WT (white bars) or ATF3−/− (black bars) fMLP-stimulated neutrophils. Representative of 2 independent experiments; N = 52 to 83 polarized, nucleus-containing neutrophils/genotype. (D) The total area of F-actin polymerization quantified from A for WT (white bars) or ATF3−/− (black bars) neutrophils stimulated by fMLP on (left) fibrinogen-coated or (right) uncoated glass slides. Representative of 3 independent experiments; N = 64 to 156 cells counted/genotype. Statistics are unpaired 2-tailed Student t test. Data are mean ± SEM. **P < .01, ***P < .001.
ATF3−/−neutrophils lacking TIAM2 exhibit dysregulated integrin-dependent cytoskeletal organization and adhesive structure regulation. (A) (Upper) WT or (lower) ATF3−/− neutrophils were allowed to adhere to uncoated glass slides and then stimulated with 10 μM fMLP and stained for polymerized F-actin by phalloidin-rhodamine and focal complexes (arrows) and focal contacts (arrowheads) by antivinculin-Alexa Fluor 488. Representative focal complex/contacts (green), polymerized F-actin (red), 4′6 diamidino-2-phenylindole–stained nuclei (blue), and merged immunofluorescent images are shown. Original magnification, ×630. Representative of 2 (vinculin) or 5 (phalloidin) independent experiments. Fluorescence images were captured at room temperature using a Leica DMI6000 fluorescence microscope at 63×/1.3 NA objective with an ORCA-ER C4742-95 camera driven by Openlab (version 5.5.0) software. Total (B) area and (C) numbers of vinculin-containing focal structures quantified from A for WT (white bars) or ATF3−/− (black bars) fMLP-stimulated neutrophils. Representative of 2 independent experiments; N = 52 to 83 polarized, nucleus-containing neutrophils/genotype. (D) The total area of F-actin polymerization quantified from A for WT (white bars) or ATF3−/− (black bars) neutrophils stimulated by fMLP on (left) fibrinogen-coated or (right) uncoated glass slides. Representative of 3 independent experiments; N = 64 to 156 cells counted/genotype. Statistics are unpaired 2-tailed Student t test. Data are mean ± SEM. **P < .01, ***P < .001.
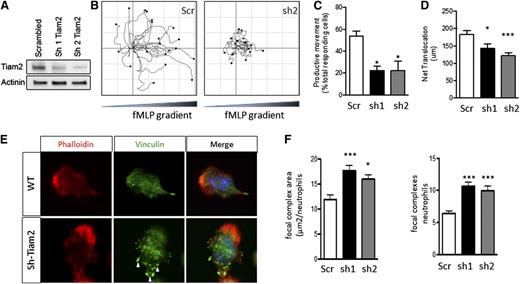
TIAM2 knockdown in WT neutrophils phenocopies the migration defects of ATF3−/− neutrophils
To define a mechanistic role for decreased TIAM2 expressed in the decreased migration and increased adhesion structures observed in ATF3−/− neutrophils, we knocked down TIAM2 expression in WT neutrophils (TIAM2KD). Two different TIAM2 shRNA constructs produced significant reductions in TIAM2 protein levels in TIAM2KD neutrophils compared with scrambled shRNA control (TIAM2cntrl) neutrophils (Figure 7A). TIAM2KD neutrophils demonstrated significant reductions in frequency of productive movement compared with TIAM2cntrl neutrophils (Figure 7B-C; supplemental Movies 3-5). TIAM2cntrl neutrophils were able to efficiently move up chemoattractant gradients, whereas TIAM2KD neutrophils were not (Figure 7B). Further recapitulating the ATF3−/− phenotype, motile TIAM2KD neutrophils demonstrated a significant reduction in net translocation in comparison with TIAM2cntrl neutrophils (Figure 7D), along with a significant increase in adhesion structure area and focal complex numbers (Figure 7E-F). Taken together with the observed inhibition of CXCL1 production in response to low-dose LPS challenge, these data identify ATF3 as a complex regulator of neutrophil recruitment and chemotaxis.
Tiam2 knockdown reproduces the ATF3−/−neutrophil phenotype in WT neutrophils. Tiam2 knockdown reproduces the ATF3−/− neutrophil phenotype in WT neutrophils. (A) TIAM2 shRNA knockdown with sequence 1 (sh1) or 2 (sh2) results in a significant reduction in TIAM2 protein expression by western blot in WT neutrophils compared with scrambled shRNA knockdown control. (B-D) Neutrophils transduced with scr (white), sh1 (black), or sh2 (gray) shRNA were placed in a Zigmond slide with chemoattractant gradient (10 nM fMLP) established to the right. Images were captured using a Zeiss Axiovert 200 microscope at 10×/0.3 NA objective, equipped with an ORCA-ER C4742-95 camera driven by Openlab, version 5.5.0 software. (B) Individual paths were traced using Image J software. Cells generating (C) productive (>20 μm) movement and (D) net distance traveled in microns were quantified. (E-F) Transduced neutrophils as in C were allowed to adhere to uncoated glass slides and then stimulated with 10 μM fMLP. Focal complexes and contacts were determined by vinculin-Alexa-fluor 488 stain as in Figure 6. Fluorescence images were captured at room temperature using a Leica DMI6000 fluorescence microscope at 63×/1.3 NA objective with an ORCA-ER C4742-95 camera driven by Openlab, version 5.5.0 software. Representative images in E and focal complex quantification in F. Results are from independent experiments: (C) N = 2 to 4 independent acquisitions, (D) N = 13 to 22 cells counted/genotype, and (E-F) N = 38 to 60 cells analyzed/genotype. Statistics are 1-way ANOVA with asterisks indicating differences compared with scr control using Dunnett’s multiple comparison post-test. Data are mean ± SEM. *P < .05, ***P < .001.
Tiam2 knockdown reproduces the ATF3−/−neutrophil phenotype in WT neutrophils. Tiam2 knockdown reproduces the ATF3−/− neutrophil phenotype in WT neutrophils. (A) TIAM2 shRNA knockdown with sequence 1 (sh1) or 2 (sh2) results in a significant reduction in TIAM2 protein expression by western blot in WT neutrophils compared with scrambled shRNA knockdown control. (B-D) Neutrophils transduced with scr (white), sh1 (black), or sh2 (gray) shRNA were placed in a Zigmond slide with chemoattractant gradient (10 nM fMLP) established to the right. Images were captured using a Zeiss Axiovert 200 microscope at 10×/0.3 NA objective, equipped with an ORCA-ER C4742-95 camera driven by Openlab, version 5.5.0 software. (B) Individual paths were traced using Image J software. Cells generating (C) productive (>20 μm) movement and (D) net distance traveled in microns were quantified. (E-F) Transduced neutrophils as in C were allowed to adhere to uncoated glass slides and then stimulated with 10 μM fMLP. Focal complexes and contacts were determined by vinculin-Alexa-fluor 488 stain as in Figure 6. Fluorescence images were captured at room temperature using a Leica DMI6000 fluorescence microscope at 63×/1.3 NA objective with an ORCA-ER C4742-95 camera driven by Openlab, version 5.5.0 software. Representative images in E and focal complex quantification in F. Results are from independent experiments: (C) N = 2 to 4 independent acquisitions, (D) N = 13 to 22 cells counted/genotype, and (E-F) N = 38 to 60 cells analyzed/genotype. Statistics are 1-way ANOVA with asterisks indicating differences compared with scr control using Dunnett’s multiple comparison post-test. Data are mean ± SEM. *P < .05, ***P < .001.
Discussion
ATF3 is a counter-regulatory immune TF1,2 that acts as an immunologic rheostat, restraining immune activation in response to transient or low levels of stimulation, yet allowing immune activation by persistent or high doses of stimulation.2,4 Here we report that ATF3 plays an unexpectedly complex role in acute neutrophilic lung inflammation. On the one hand, ATF3 inhibits LPS-dependent CXCL1 production by lung epithelia, which would tend to reduce neutrophil recruitment. On the other hand, neutrophils require ATF3 expression to migrate normally, likely through developmental regulation of TIAM2 expression, which would tend to facilitate neutrophil recruitment.
These studies are the first to demonstrate regulation of epithelial Cxcl1 expression by ATF3. We identified an ATF3 binding site unique to the Cxcl1 promoter among ELR+ chemokines and demonstrated dose-dependent LPS-driven CXCL1 transcript and protein levels in ATF3−/− airways and pulmonary epithelium. Specific regulation is likely due to direct Cxcl1 promoter occupancy by ATF3, as others have immunoprecipitated ATF3 at the CXCL1 promoter following ectopic overexpression in human cancer cell lines.33 A previous study reported increased (mRNA) expression of the entire ELR+ chemokine family in ATF3−/− lung homogenates in an allergic asthma model, including members lacking consensus ATF3 binding sites.34 We demonstrated ATF3-specific, LPS-dependent overexpression of CXCL1 mRNA and protein in airways and lung epithelia within 24 hours of challenge, whereas Gilchrist et al35 assayed transcripts in lung homogenates following a 4-week sensitization/challenge model. Therefore, their observation of globally increased ELR+ family expression in ATF3−/− lungs likely reflects global perturbations in the immune response in asthma.
In the airway, the kinetics of LPS-induced CXCL1 production and neutrophil recruitment did not overlap, although CXCL1 was the primary driver of neutrophil recruitment, as indicated by CXCL1 neutralization studies. The mechanism underlying these events is unclear. Unique among ELR+ chemokines, airway-produced CXCL1 is actively transcytosed into the systemic circulation.19 Therefore, systemic CXCL1 could be the inciting event responsible for neutrophil recruitment by priming circulating neutrophils and inducing extravasation into the lung interstitium, whereas other molecules (potentially CXCL2 and/or CXCL5) contribute to the continued accumulation of neutrophils in the alveolar spaces independent of or in conjunction with CXCL1. This also suggests that increases in CXCL1 concentrations in BALF may only represent a portion of the dysregulated CXCL1 response in ATF3−/− mice. Therefore, future kinetic experiments evaluating neutrophil accumulation in the pulmonary circulation, interstitium, and alveolar spaces,36 as well as definition of systemic ELR+ chemokine production, is warranted. Nevertheless, the current studies uncover a fundamental role for ATF3-mediated regulation of CXCL1 in acute lung inflammation.
The most surprising result of our studies was the lack of increased ATF3−/− neutrophil recruitment to the lungs of ATF3−/− mice, despite significant increases in CXCL1. Time-lapse microscopy of chemotaxing neutrophils demonstrated a remarkable ATF3−/− neutrophil-intrinsic migration defect. Neutrophil migration necessitates rapid cell body reorganization and is commonly coordinated by signaling molecules leading to rearrangement of diverse cytoskeletal structures. TFs are more likely involved in target gene regulation, which occurs on time scales significantly longer than the immediate responses necessitated by cellular migration. Therefore, this observation raised certain questions. (1) Does ATF3 regulate neutrophil development by modulating the expression of genes necessary for proper mature neutrophil function? (2) Which proteins involved in neutrophil chemotaxis are regulated by ATF3 during neutrophil development? (3) What is/are the mechanism(s) underlying ATF3−/− neutrophil-intrinsic chemotaxis defects?
Our data demonstrate that ATF3 is critical for generating fully functional mature neutrophils, likely by developmental regulation of effector proteins necessary for migration, such as TIAM2. Although ATF3 expression is barely detectable in neutrophils, it is necessary for TIAM2 expression during neutrophil development. Evidence for a regulatory role for ATF3 in neutrophil development has been described.37 JDP2 is an important negative regulator of ATF3 expression37-39 whose depletion results in elevated Atf3 expression, decreased Ly6G expression (a neutrophil maturation marker), and defective effector function (eg, superoxide production, neutrophil extracellular trap formation).37 Ectopic ATF3 overexpression throughout neutrophil differentiation recapitulated the immature phenotype of JDP2−/− neutrophils.37 When considered in light of our findings, however, these data imply that ATF3 induction and subsequent down-modulation may be necessary for terminal neutrophil development. Similar to the waves of TFs integral to neutrophil development (ie, PU.1, C/EBP-α, Gfi-1, and C/EBP-ε), ATF3 induction may modulate subsequent waves of TF expression,9 followed by ATF3 silencing necessary to release gene targets from transcriptional repression. These issues clearly deserve further exploration.
There are numerous genes integral to chemotaxis that could contribute to the observed chemotaxis defect in ATF3−/− mice. However, the time scale of chemotaxis likely precludes de novo transcriptional regulation of these potential genes by ATF3. Nonetheless, the protein products of genes Pik3cg, Rac1, and Rac2 participate in many of the processes observed to be defective in ATF3−/− neutrophil chemotaxis. On further evaluation (by microarray analysis and qRT-PCR), however, none of these genes were dysregulated in ATF3−/− neutrophils, notably despite the presence of a putative ATF3 binding site in the promoters of Pik3cg and Rac2.
TIAM2 is a specific Rac1-activating guanine exchange factor40,41 that controls cellular motility by promoting FA disassembly in epithelial cells.21 We observed significantly increased focal complex areas and numbers in ATF3−/− neutrophils, associated with an absence of Tiam2 expression. Epithelial TIAM2 knockdown resulted in reduced rates of FA disassembly and consequently enlarged FAs and reduced migratory speeds.21 Therefore, TIAM2-mediated dysregulated focal complex disassembly is a plausible molecular mechanism for reduced ATF3−/− neutrophil chemotaxis. This hypothesis was further supported by lentivirus-mediated shRNA knockdown of TIAM2 in ATF3-sufficient WT neutrophils, which resulted in a phenocopy of the ATF3−/− neutrophil chemotaxis phenotype. Interestingly, ATF3 overexpression in breast cancer cell lines up-regulates genes involved in cancer metastasis42 and confers metastatic potential to prostate cancer cell lines.43 Thus, ATF3-dependent migration effects may not be restricted to neutrophils and may represent a broader ATF3 modulation of cellular adhesion.
In addition to increased focal complexes, we observed striking increases in F-actin polymerization in ATF3−/− neutrophils. It is unclear whether this is secondary to dysregulated focal complex disassembly or represents an additional ATF3-dependent effect. In the migratory cycle, lamellipodia are stabilized to the substratum via adhesion, enabling cell body translocation and efficient migration. These adhesion points are then dissembled to complete movement. It is possible that increased adhesion complexes cause abnormal enlargement of the lamellipodia. Alternatively, Rac1−/− neutrophils display increased polymerized F-actin and aberrant lamellipodia morphology in response to fMLP stimulation,26 similar to that seen in ATF3−/− neutrophils. Although the TIAM2-mediated effect on epithelial FAs was shown to be Rac1 dependent,21 focal complexes were not evaluated in Rac1−/− neutrophils.26 Therefore, ATF3-dependent Tiam2 regulation may have multifaceted effects on neutrophil chemotaxis.
Overall, our study describes novel complex roles for ATF3 in neutrophil recruitment: inhibition of epithelial CXCL1 production in addition to neutrophil-intrinsic promotion of chemotaxis, likely through developmental modulation of TIAM2 expression and focal complex and cytoskeletal regulation. Increased understanding of the developmental and regulatory ATF3 effects on neutrophil recruitment may identify novel therapeutic targets in disease marked by aberrant neutrophilic inflammation.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Cheryl Minges and Maria Holdcroft for technical help, the Cincinnati Children’s Hospital Medical Center animal husbandry staff, and Jeffery Bailey and Victoria Summey for help with BM transplantation studies.
This work was supported by National Institutes of Health, National Heart, Lung and Blood Institute grants R01 HL094576 (to C.L.K.) and R01 HL090676 (to M.-D.F.); and National Institute of General Medical Sciences grant T-32 GM063483 (Medical Scientist Training Program) and Cystic Fibrosis Foundation grant R457CR11 (to C.L.K.).
Authorship
Contribution: N.D.B., M.-D.F., and C.L.K. designed the experiments, interpreted the results, and drafted the paper; H.L.G. and K.H. aided in data interpretation and experimental design; N.D.B. performed the research with help from M.-D.F., S.K., and J.W.M.; J.D.P. provided expertise on microarray analysis; and T.H. provided ATF3−/− mice and aided in editing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for C.L.K. is The Bill & Melinda Gates Foundation, Seattle, WA.
Correspondence: Marie-Dominique Filippi, Experimental Hematology and Cancer Biology, CCHMC, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: marie-dominique.filippi@cchmc.org; and Christopher L. Karp, Bill & Melinda Gates Foundation, 500 Fifth Ave North, Seattle, WA 98109; e-mail: chris.karp@cchmc.org.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal