In this issue of Blood, Allinne et al propose the nucleolin-dependent activation of the translocated CCND1 allele in mantle cell lymphoma (MCL) because of its relocalization to a transcriptionally favorable area in the perinucleolar region.1
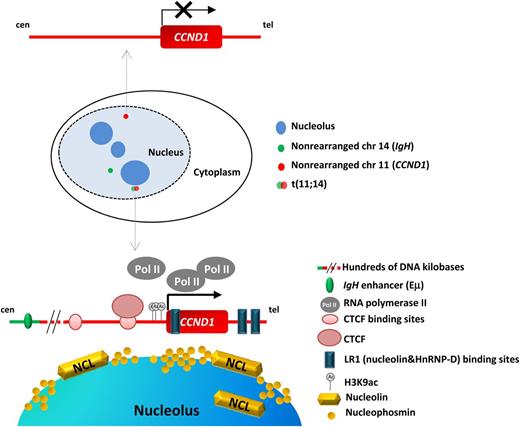
Model for CCND1 transcription activation proposed by Allinne and colleagues. In the schematic representation of the nucleus, the wild-type CCND1 is indicated in red and the IgH in green, whereas the der(14) corresponding to the translocated IgH-CCND1 allele (fusion of red and green signals) is located in the periphery of a nucleolus (dark blue). The t(11;14) shifts toward the nucleolus. The intact CCND1 allele, transcriptionally inactive, is represented in the upper part of the figure; the translocated CCND1 allele, transcriptionally active, is represented in the lower part. Several regions related with its activation are highlighted: the CCND1-binding sites for CTCF and LR1, the active acetylation marks in the promoter and the nucleolus close proximity, with abundant nucleolin, active PolII clusters and CTCF. Distant Eμ IgH enhancer is also indicated. cen, centromere; tel, telomere.
Model for CCND1 transcription activation proposed by Allinne and colleagues. In the schematic representation of the nucleus, the wild-type CCND1 is indicated in red and the IgH in green, whereas the der(14) corresponding to the translocated IgH-CCND1 allele (fusion of red and green signals) is located in the periphery of a nucleolus (dark blue). The t(11;14) shifts toward the nucleolus. The intact CCND1 allele, transcriptionally inactive, is represented in the upper part of the figure; the translocated CCND1 allele, transcriptionally active, is represented in the lower part. Several regions related with its activation are highlighted: the CCND1-binding sites for CTCF and LR1, the active acetylation marks in the promoter and the nucleolus close proximity, with abundant nucleolin, active PolII clusters and CTCF. Distant Eμ IgH enhancer is also indicated. cen, centromere; tel, telomere.
MCL is a CD5-positive mature B-cell neoplasm characterized by the t(11;14)(q13;q32) translocation leading to overexpression of cyclin D1, which is not expressed in normal B lymphocytes.2 This hallmark translocation is considered to be the primary genetic alteration of MCL and is found in virtually all cases. Although it may be an essential hit for the initiation of tumor development, it does not seem sufficient. This is mainly because of the weak oncogenicity of cyclin D1, which requires the cooperation of other genetic events to fully transform the lymphoid cells.3 Several molecular studies have identified additional alterations in components of cell cycle regulation, DNA damage response, cell survival pathways, NOTCH and nuclear factor-κB pathways, and chromatin modification machinery.2,4,5 These secondary molecular alterations are frequently associated with chromosomal gains and losses that target important genes as well as with an accumulation of somatic mutations. During the past 2 decades, much effort has been spent on deciphering the spectrum of these secondary genetic alterations in MCL.2 However, little is known about the molecular causes of the transcriptional activation of CCND1 in MCL cells.
In MCL, the t(11;14) seems to occur in pre-B lymphocytes of the bone marrow. The consequence of this rearrangement is that 1 CCND1 allele (located at 11q13) becomes juxtaposed to the potent intronic enhancer (Eμ) of IgH (located at 14q32). Although the distance between the regulatory Eμ element and the CCND1 gene is several hundred kilobases, it is believed that the mechanism responsible for the transcriptional activation of CCND1 is due to the action of this potent IgH enhancer.
The first studies on the transcriptional activation of the CCND1-translocated allele observed DNA hypomethylation, histone hyperacetylation, and especially binding of RNA pol II (PolII) to the CCND1 promoter region and to the regulatory IgH region, as detected by chromatin immunoprecipitation (ChIP).6 Noteworthy is that the same group provided additional clues for the CCND1 activation in MCL cells, suggesting a functional role for CCCTC binding factor (CTCF) and nucleophosmin (NPM).7 In cells carrying the t(11;14), CTCF and NPM were both associated with the IgH regulatory region and the CCND1 promoter, suggesting that CTCF could play a role in tethering both Eμ and the CCND1 promoter in the nucleolar periphery through its interaction with NPM.
With these previous observations in mind and with the knowledge that in the nucleus the chromosomes occupy specific regions or territories as well as that gross genetic alterations and translocations may cause changes in the nuclear localization, Allinne et al1 have gone a step further. The authors have investigated the changes in the nuclear localization of both the intact and rearranged CCND1 alleles in MCL with the aim of determining the molecular mechanism triggering CCND1 activation in the translocated allele. They carefully measured the position and distance of the translocated and nontranslocated CCND1 alleles by 3-dimensional fluorescence in situ hybridization and observed that the translocated allele is positioned within a perinucleolar area, whereas both the normal IgH and CCDN1 loci lay further away from the nucleolus than their translocated counterparts (see figure). The relocalization of the translocated CCND1 allele occurs in a nuclear region with high concentrations of active PolII molecules and a potent transcriptional activator, LR1, formed by heterodimers of nucleolin and HnRNP-D. Nucleolin is very abundant in the nucleolus and, among other functions, is implicated in transcriptional regulation. In contrast, the nontranslocated CCND1 allele did not colocalize with PolII transcription clusters.
Interestingly, of the 2 putative CTCF-binding sites near CCND1, the closest to the gene is highly occupied by CTCF in MCL cell lines compared with normal lymphocytes. This finding suggests that upstream enhancer elements might not play a role in CCDN1 activation because they may be blocked by the CTCF-bound insulator. Furthermore, the authors propose that CTCF itself may play a role in CCDN1 activation, although they did not provide further evidence supporting this interesting hypothesis. Elegantly, the authors show that there are 3 LR1-binding sites in CCND1 that have enhancer activity. Thus, the high concentration of LR1 and its binding to the LR1 sites of CCND1 might be the reason CCDN1 is activated in these cells. Additionally, as a proof of concept, the authors performed ChIP-on-chip experiments with an antinucleolin antibody and confirmed the interactions of CyclinD1 protein and nucleolin. Furthermore, this experiment also revealed a high concentration of lysine 9–acetylated histone H3 at the CCND1 promoter, an expected result confirming that the translocated CCND1 allele in these cells is transcriptionally active (see figure). In addition, the treatment of the MCL cell lines with a pan-histone deacetylase inhibitor perturbed the overall nuclear structure and dramatically reduced the transcription levels of CCND1.
The authors also extended the relocalization studies of the translocated allele to MYC translocation in an endemic Burkitt lymphoma cell line with the t(8;14) translocation. By 3-dimensional fluorescence in situ hybridization, they found a similar relocalization of rearranged alleles to the perinucleolar territories, suggesting that similar transcriptional mechanisms may lead to the activation of MYC in this translocation. In fact, nucleolin seemed to be able to activate the transcription of MYC more efficiently than in the case of CCND1.
Overall, these observations shed light on the mechanisms leading to transcriptional deregulation in common chromosomal oncogenic translocations in B-cell lymphomas being the translocated allele activated by its relocalization to nucleolin-rich regions of the nucleus.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal