Key Points
The x-ray crystal structure of the N2 domain from HRG at 1.93 Å resolution is presented.
The structure reveals an S-glutathionyl adduct at Cys185, which has implications for angiogenic regulation.
Abstract
Histidine-rich glycoprotein (HRG) is a plasma protein consisting of 6 distinct functional domains and is an important regulator of key cardiovascular processes, including angiogenesis and coagulation. The protein is composed of 2 N-terminal domains (N1 and N2), 2 proline-rich regions (PRR1 and PRR2) that flank a histidine-rich region (HRR), and a C-terminal domain. To date, structural information of HRG has largely come from sequence analysis and spectroscopic studies. It is thought that an HRG fragment containing the HRR, released via plasmin-mediated cleavage, acts as a negative regulator of angiogenesis in vivo. However, its release also requires cleavage of a disulphide bond suggesting that its activity is mediated by a redox process. Here, we present a 1.93 Å resolution crystal structure of the N2 domain of serum-purified rabbit HRG. The structure confirms that the N2 domain, which along with the N1 domain, forms an important molecular interaction site on HRG, possesses a cystatin-like fold composed of a 5-stranded antiparallel β-sheet wrapped around a 5-turn α-helix. A native N-linked glycosylation site was identified at Asn184. Moreover, the structure reveals the presence of an S-glutathionyl adduct at Cys185, which has implications for the redox-mediated release of the antiangiogenic cleavage product from HRG.
Introduction
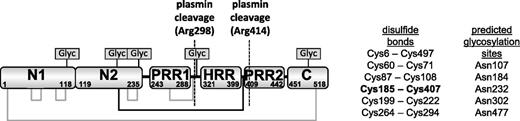
Histidine-rich glycoprotein (HRG) is a plasma protein that regulates angiogenesis, coagulation, and immune function in vertebrates. Human HRG has an approximate mass of 70 kDa and is present in plasma at low micromolar concentrations (ca ∼1.5 μM). The protein is arranged into 6 domains: 2 N-terminal domains (N1 and N2), a central histidine-rich region (HRR) flanked by 2 proline rich regions (PRR1 and PRR2), and a C-terminal domain (C). The N1, N2, and C domains are highly conserved between species, as is an arrangement of 6 disulfide bridges (Figure 1). In plasma, HRG binds to and regulates the function of a diverse variety of targets that include fibrinogen, plasminogen, thrombospondin, immunoglobulin G (IgG), complement factors, and heparin as well as cell-surface molecules such as Fcγ receptors and heparan sulfate.1-9 HRG binds divalent metal cations within the HRR.10 In particular, Zn2+ is known to bind this region and can modulate HRG activity by altering the protein’s affinity for other targets.11
Domain structure of rabbit HRG showing the disulfide bridging arrangement and 5 putative glycosylation sites. The disulfide bridge between the N2 domain and the HRR/PRR region is highlighted in bold and by the black lines.
Domain structure of rabbit HRG showing the disulfide bridging arrangement and 5 putative glycosylation sites. The disulfide bridge between the N2 domain and the HRR/PRR region is highlighted in bold and by the black lines.
HRG is heavily glycosylated; the human protein has 6 putative N-linked glycosylation sites.11 The histidine- and proline-rich regions are predicted to be intrinsically disordered but N1, N2, and the C-terminal domains are likely to have ordered structures. The N1 and N2 domains share a high degree of sequence similarity to members of the cystatin superfamily of cysteine protease inhibitors.12 Type 1 cystatins (also known as stefins) are characterized by a lack of disulfide bonds, whereas the type 2 cystatins have 2 disulfide bonds.13 These groups include the proteins cystatin B and cystatin C, respectively, which have proposed roles in the inhibition of tumor neovascularization.14 The similarity between cystatins and N1 and N2 domain sequences has led to the classification of HRG as a type 3 cystatin, along with fetuins and kininogens.15 However, it has been shown that HRG is unable to inhibit cysteine peptidases of the papain (C1) family suggesting that it does not possess cysteine protease inhibitor activity.15 The N-terminal domains of HRG also bear similarity to the N-terminal region of antithrombin III, where the respective domains of both proteins are implicated in heparin binding.16 This and the fact that both HRG and antithrombin III regulate thrombin activity by competing with one another for binding to heparin17 points toward an evolutionary relationship between the 2 proteins. The N1 and N2 terminal domains of HRG are also implicated in the control of immune functioning through interactions with C1q, IgG, and FcγRI.4,9
HRG regulates the formation of new blood vessels in both a pro-and antiangiogenic manner. The proangiogenic activity of HRG stems from its high affinity toward thrombospondin, an inhibitor of angiogenesis, via CD36/LIMP-2/EMP structural homology (CLESH) motifs contained within its C-terminal domain.18 A similar proangiogenic mechanism is thought to occur through its binding of vasculostatin.19 An antiangiogenic effect mediated by HRG was localized to the HRR by studies using proteolytic fragments, truncated HRG proteins, and peptides based on the HRR.20 HRG treated with the plasma protease, plasmin, inhibits the proliferation of endothelial cells both in vitro and in vivo.20 It is known that some physiological functions of HRG require plasmin-mediated release of a fragment from the intact protein that contains the HRR and part of PRR2 (HRR/PRR). Such functions include its antimicrobial and endotoxin-neutralizing properties.21,22 Recombinant HRG proteins/fragments of varying truncations have been used to study the antiangiogenic properties of HRG in fibrosarcoma tumor model mice.23 HRG proteins that contained the HRR reduced tumor growth and vascularization, whereas those lacking this domain had no effect. These data strongly suggest that the HRR/PRR fragment is the inhibitor of angiogenesis. A 35-amino acid peptide, HRGP330, based upon a sequence within the HRR has been identified as the minimal active domain with antiangiogenic properties in vitro and in vivo.24,25 This peptide acts by disrupting integrin-linked kinase and focal adhesion kinase (FAK) functions, consequently leading to an arrest in endothelial cell motility.22 Collectively, these studies implicate HRG in the control of angiogenesis through release of HRR/PRR via a proteolytically regulated mechanism.26,27
Plasminogen (the precursor to plasmin) is closely associated with HRG, and it is estimated that 50% of plasminogen in plasma is bound to HRG.1 Proteolysis by plasmin alone is not enough to release HRR/PRR due to the presence of a particular disulfide bond within HRG. The HRG structure is held together by an arrangement of 6 disulfide bonds, 4 of which are intradomain and 2 are interdomain. An interdomain disulfide bond connects the N1 and C-terminal domains, similar to a disulfide bridge that exists in other members of the type 3 cystatin family. The other interdomain disulfide is unique to HRG and connects the N2 domain to a linker region between the HRR and PRR2 domains (Figure 1).28 A peptide corresponding to the HRR/PRR fragment has been identified in human tissue samples but the mechanism by which the interdomain disulfide is reduced is not known.29
Using x-ray crystallography we present the first structural characterization of HRG. We reveal that the N2 domain of serum-purified rabbit HRG does indeed possess a cystatin-like fold and is N-glycosylated at Asn184. We also observed a glutathione (GSH) adduct at Cys185. This cysteine residue is known to form a disulfide bridge with Cys407 that links the N2 and HRR/PRR-associated regions in the intact protein. Because no reducing or proteolytic conditions were used during isolation or crystallization, these data strongly suggest that GSH (together with plasmin) can act as an angiogenic regulator by controlling plasmin-mediated release of the HRR/PRR fragment in vivo.
Materials and methods
Purification and crystallization of rabbit HRG
Rabbit HRG was purified directly from rabbit serum or plasma using nickel-affinity chromatography. Rabbit serum/plasma was filtered using a 0.45 µM filter (Sartorius) and applied to a HisTrap HP column (GE Healthcare Life Sciences) preequilibrated with 50 mM Tris, 150 mM NaCl, 5 mM imidazole at pH 8. The column was then washed using the same buffer, before HRG was eluted using the same buffer supplemented with 400 mM imidazole. Prior to crystal trials, the serum-prepared rabbit HRG was further purified by size exclusion chromatography, using an S-200 column (GE Healthcare Life Sciences) preequilibrated in 10 mM Tris, 150 mM NaCl, pH 8. HRG eluted in a single symmetrical peak consistent with a molecular weight of ∼140 kDa. HRG homogeneity was assayed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and fractions containing HRG were pooled and concentrated to 16 mg/mL. HRG was screened for crystallization using 2 commercial crystal screens, JCSG+, and PACT (Qiagen), alongside in-house stochastically designed screens on an Art Robbins Gryphon crystallization robot by sitting-drop vapor diffusion. Two visibly distinct crystal morphologies were apparent after 4 weeks. Crystals that displayed a cubic morphology only diffracted to 5.2 Å, however, rod-shaped crystals grown from 22.5% polyethylene glycol (PEG) modified monomethyl ether (MME) 5000, 0.073 M potassium sodium tartrate, and 0.1 M 4-morpholineethanesulfonic acid (MES), pH 6.5, diffracted considerably better, and subsequently both native and heavy atom soaked data sets were collected from this morphology of crystal. For phasing, a rod-shaped crystal was harvested and soaked for 5 minutes in mother liquor doped with 50 mM K2PtCl4 before being back-soaked in cryoprotectant (mother liquor with 25% glycerol) for 10 seconds and immediately cryocooled in liquid nitrogen. Data were collected on Diamond beamline I02 as 1000 nonoverlapping images using a Pilatus 6M detector. At a later date, a high-resolution native data set was collected from a different rod-shaped crystal on Diamond beamline I04-1. All data were processed with xia2.30
The data obtained from crystals soaked with K2PtCl4 were analyzed using the PHENIX program package.31 Autosol, using both anomalous and isomorphous difference, identified 2 Pt sites in space group P3121 with a figure of merit of 0.18 at 3.1 Å. Density modification and extension to 2.9 Å using Autobuild gave a preliminary model of 107 residues and an R-factor of 30%. This preliminary model was rebuilt by manual inspection using COOT.32 Data completeness at 2.1 Å is 96% and only dropped below 90% at a resolution of 2.0 Å (completeness 83%). Refinement to 2.1 Å yields an R = 18.3% and Rf = 22.2%. We decided to include all data to 1.93 Å where I/σ (I) = 2.1, completeness = 69.2%, and Rmerge = 33% for the highest resolution shell of data (supplemental Table 1, available on the Blood Web site). Data collection and structure refinement statistics are presented in Table 1. The final structure was deposited in the RCSB Protein Databank (www.rcsb.org/; PDB code: 4CCV).
Crystallographic data collection and structure refinement statistics
| . | K2PtCl4-soaked . | Native . |
|---|---|---|
| Data collection | ||
| Beamline | Diamond I02 | Diamond I04-1 |
| Wavelength, Å | 0.97 | 0.92 |
| Space group | P 31 2 1 | P 31 2 1 |
| Cell dimensions, Å, ° | 77.6, 77.6, 69.2 | 77.1, 77.1, 69.2 |
| 90, 90, 120 | 90, 90, 120 | |
| Resolution, Å(high resolution) | 69.2-2.91(2.99-2.91) | 34.7-1.93(2.00-1.93) |
| Rmerge | 10.0 (92.3) | 5.1 (33.0) |
| I/σ (I) | 29.2 (5.2) | 11.3 (2.1) |
| Completeness (%) | 99.5 (99.2) | 93.7 (69.2) |
| Average redundancy | 26.9 (28.6) | 4.1 (2.7) |
| Vm, Å3/Da | 4.3 | 4.2 |
| Solvent, % | 71.4 | 70.9 |
| Refinement | ||
| Unique reflections | 17163 | |
| Rwork/Rfree | 0.18/0.22 | |
| Geometric deviations | ||
| Bonds, Å/Angles, ° | 0.017/2.02 | |
| No. of atoms | 1104 | |
| Protein | 952 | |
| Water | 75 | |
| Sugar | 39 | |
| GSH | 20 | |
| Glycerol | 18 | |
| B factors, Å | ||
| Protein | 41 | |
| Water | 51.8 | |
| Sugar | 69.6 | |
| GSH | 63.6 | |
| Glycerol | 74.3 | |
| Ramachandran | ||
| Allowed/disallowed, % | 100/0 | |
| Molprobity score/centile | 3.52/100 | |
| PDB code | 4CCV |
| . | K2PtCl4-soaked . | Native . |
|---|---|---|
| Data collection | ||
| Beamline | Diamond I02 | Diamond I04-1 |
| Wavelength, Å | 0.97 | 0.92 |
| Space group | P 31 2 1 | P 31 2 1 |
| Cell dimensions, Å, ° | 77.6, 77.6, 69.2 | 77.1, 77.1, 69.2 |
| 90, 90, 120 | 90, 90, 120 | |
| Resolution, Å(high resolution) | 69.2-2.91(2.99-2.91) | 34.7-1.93(2.00-1.93) |
| Rmerge | 10.0 (92.3) | 5.1 (33.0) |
| I/σ (I) | 29.2 (5.2) | 11.3 (2.1) |
| Completeness (%) | 99.5 (99.2) | 93.7 (69.2) |
| Average redundancy | 26.9 (28.6) | 4.1 (2.7) |
| Vm, Å3/Da | 4.3 | 4.2 |
| Solvent, % | 71.4 | 70.9 |
| Refinement | ||
| Unique reflections | 17163 | |
| Rwork/Rfree | 0.18/0.22 | |
| Geometric deviations | ||
| Bonds, Å/Angles, ° | 0.017/2.02 | |
| No. of atoms | 1104 | |
| Protein | 952 | |
| Water | 75 | |
| Sugar | 39 | |
| GSH | 20 | |
| Glycerol | 18 | |
| B factors, Å | ||
| Protein | 41 | |
| Water | 51.8 | |
| Sugar | 69.6 | |
| GSH | 63.6 | |
| Glycerol | 74.3 | |
| Ramachandran | ||
| Allowed/disallowed, % | 100/0 | |
| Molprobity score/centile | 3.52/100 | |
| PDB code | 4CCV |
Values in parentheses refer to the highest resolution shell.
Analysis of plasmin- and GSH-mediated cleavage products
To understand the redox state of HRG in plasma and the effect of plasmin activity, purified rabbit HRG (prepared from serum) was subjected to analysis by SDS-PAGE followed by Coomassie staining under a range of conditions. Native HRG without reducing agent, native HRG with reducing agent and plasmin-treated HRG with reducing agent were all analyzed. To further characterize HRR/PRR, a band (∼25 kDa) putatively corresponding to this fragment was cut and submitted for MS/MS analysis.
Mass spectrometry
The excised gel band was cut into 1 mm3 pieces. These were then subjected to in-gel digestion, using a ProGest Investigator in-gel digestion robot (Genomic Solutions). Briefly, the gel pieces were destained by washing with acetonitrile and subjected to reduction and alkylation before digestion with thermolysin at 55°C. The peptides were extracted with 10% formic acid. The digest solution (0.5 µL) was applied to the matrix-assisted laser desorption/ionization (MALDI) target along with α-cyano-4-hydroxycinnamic acid matrix (0.5 µL, 10 mg/mL in 50:50 acetonitrile:0.1% trifluoroacetic acid) and allowed to dry.
MALDI mass spectrometry (MS) was acquired using a 4800 MALDI time of flight (TOF) Analyzer (AB Sciex) equipped with a Nd:YAG 355 nm laser and calibrated using a mixture of peptides. The spot was initially analyzed in positive MS mode between 800 and 4000 m/z, by averaging 1000 laser spots. The most intense peptides (up to 15) were selected for MS/MS analysis and acquired to a maximum of 3000 laser shots or until the accumulated spectrum reached a S:N ratio of 35. All MS/MS data were acquired using a collision energy of 1 keV. The MS/MS data were analyzed, using the ProteinPilot 4.1 Paragon algorithm (AB Sciex) searching against an internal database to which the HRG sequence had been added, with no enzyme digestion parameters selected and carbamidomethyl modification of cysteines selected.
Results
Purification and crystallization of HRG
Because rabbit HRG is ∼10 times more abundant in rabbit plasma than human HRG is in human plasma (0.9 mg/mL cf 0.1 mg/mL),26 we proceeded with rabbit HRG to obtain viable yields for extensive crystal trials. The properties of rabbit HRG have been extensively studied and are similar to that of human HRG.22,26 Rabbit HRG was purified from rabbit serum with a yield of 80% and was of very high purity (>95%). The purified preparation was concentrated to 16 mg/mL for crystal screening. The condition from which rod-shaped crystals grew, 21.3% PEGMME 5K, 0.03M potassium sodium tartrate, 0.1 M MES pH 6.5, is from an in-house stochastically designed crystal screen. Expanded trials of this condition yielded reproducible highly diffracting rod-shaped crystals after 4 weeks’ incubation at 20°C. Crystals with a cubic morphology were also identified. However, these cubic crystals diffracted relatively poorly and proved difficult to reproduce. Subsequently, it was decided that we would concentrate our efforts on the rod-shaped crystals.
Structure of rabbit HRG N2 domain
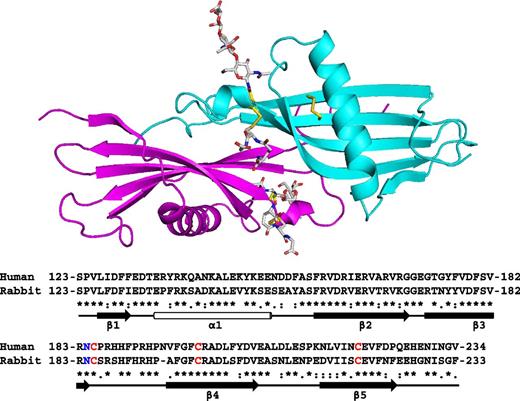
The crystal structure of the N2 domain of rabbit HRG was solved to a resolution of 1.93 Å. The structure contains 1 complete N2 domain (115 amino acids from Ser123 to Arg237). The asymmetric unit contains a single chain, however, due to the symmetry of the crystal, dimeric arrangements of the domain exists within the unit cell as shown in the upper panel of Figure 2. The biological relevance of the arrangements were evaluated by the PISA server (www.ebi.ac.uk), which predicts by analyzing the interactions between chains, whether these are likely to be stable in solution and thus meaningful. The analysis identified the arrangement shown in Figure 2 as stable (significance of 1, the maximum probability) and this arrangement buries 1659 Å2. The rabbit and human HRG proteins share 64% identity and 69% similarity overall.26 The respective N2 domain sequences of the 2 proteins are highly homologous (as can be seen in the lower panel of Figure 2; 68% identity, 93% similarity) and it is therefore likely that these share the same fold. Also critically, the cysteine residues that form the interdomain disulfide bridge linking the N2 and HRR/PRR2 regions are conserved in both human and rabbit proteins.
Crystal structure of the HRG N2 domain. The structure represents a dimeric form of the N2 domain composed of 2 identical chains, A (cyan) and B (magenta). A sequence alignment of rabbit and human N2 domains is shown with secondary structure elements highlighted; β-sheets (arrows) and the α-helix (cylinder). Conserved cysteine residues are shown in red and the N-glycosylated Asn184 residue in blue.
Crystal structure of the HRG N2 domain. The structure represents a dimeric form of the N2 domain composed of 2 identical chains, A (cyan) and B (magenta). A sequence alignment of rabbit and human N2 domains is shown with secondary structure elements highlighted; β-sheets (arrows) and the α-helix (cylinder). Conserved cysteine residues are shown in red and the N-glycosylated Asn184 residue in blue.
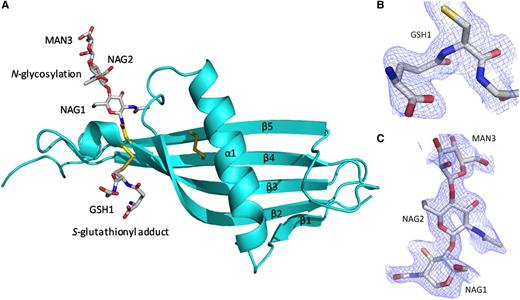
The N2 domain sequence is homologous with cystatins and HRG itself is classified as a type 3 cystatin.15 The structure confirms the N2 domain possesses a cystatin-like fold consisting of a 5-stranded antiparallel β-sheet wrapped around a 5-turn α-helix (Figure 3A). The fold starts from β1 (Leu126-Phe129) and follows through to 5-turn α-helix (Glu134-Glu150) with β2 (Phe157-Lys169), β3 (Thr174-Asn184), β4 (Ala195-Phe205), and β5 (Glu216-Asn226) wrapping and twisting around the central α-helix. In the electron density maps an S-glutathionyl adduct at Cys185 (which forms a mixed disulfide; GSH1) and an N-linked glycan at the conserved Asn184 are observed (Figure 3B-C, respectively). At each N-linked glycan chain, 2 N-acetyl glucosamine molecules (NAG1 and NAG2) and 1 α-d-mannose molecule (MAN3) can be modeled as shown.
The cystatin-like fold of the HRG N2 domain. (A) Chain A (in the same orientation as Figure 2) is shown depicting the 5 β-strands (β1-β5) wrapping and twisting around the α-helix (α1). The internal disulfide bridge can be seen linking β4 and β5. Glycosylation at Asn184 and the S-glutathionyl adduct at Cys185 are shown. The wrapping of the β-sheet (β1-β4) round the α-helix (α1) can also be observed. (B) Fo-Fc electron density map showing the S-glutathionyl adduct (GSH1) bound to Cys185 as a mixed disulfide. (C) Fo-Fc electron density map for the carbohydrate chain linked to Asn184, showing the first 3 sugars, NAG1, NAG2, and MAN3. (B-C) Fo-Fc maps (blue chicken wire contoured at 1σ, carve radius 1.6 Å) were calculated from a model that has never contained NAG, MAN, or GSH.
The cystatin-like fold of the HRG N2 domain. (A) Chain A (in the same orientation as Figure 2) is shown depicting the 5 β-strands (β1-β5) wrapping and twisting around the α-helix (α1). The internal disulfide bridge can be seen linking β4 and β5. Glycosylation at Asn184 and the S-glutathionyl adduct at Cys185 are shown. The wrapping of the β-sheet (β1-β4) round the α-helix (α1) can also be observed. (B) Fo-Fc electron density map showing the S-glutathionyl adduct (GSH1) bound to Cys185 as a mixed disulfide. (C) Fo-Fc electron density map for the carbohydrate chain linked to Asn184, showing the first 3 sugars, NAG1, NAG2, and MAN3. (B-C) Fo-Fc maps (blue chicken wire contoured at 1σ, carve radius 1.6 Å) were calculated from a model that has never contained NAG, MAN, or GSH.
Identification and characterization of HRR/PRR
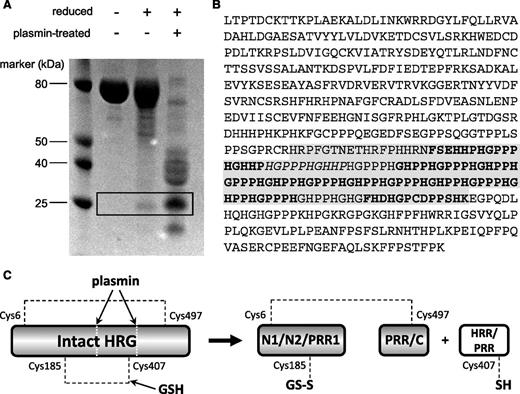
To identify the HRR/PRR fragment, purified rabbit HRG was either left untreated, reduced with dithiothreitol (DTT), or reduced and subjected to plasmin cleavage. The resultant samples were then analyzed by SDS-PAGE (Figure 4A). The gel pattern shows interesting features when comparing the appearance of the protein under the range of conditions tested. Under nonreducing conditions, the native protein runs essentially as a single band, which is expected because no proteolytic enzyme was used. Although a proportion of the protein is cleaved by plasmin in circulation the disulfide bridging (most notably the Cys185-Cys407 bond) holds the protein together. When purified HRG is reduced by DTT, the majority of the protein sample appears unaltered, but a band of ∼25 kDa can be observed. This band was cut from the gel, and the protein reduced and alkylated and digested with thermolysin and the peptides analyzed by MALDI MS and MS/MS. Peptide fragments were identified (as shown in Figure 4B) that confirmed this band to correspond to the HRR/PRR fragment. The fragments identified included a peptide containing Cys407, which forms a disulfide bridge with Cys185 in the nonreduced protein. The release of HRR/PRR following treatment of purified HRG with DTT can be explained by in vivo plasmin activity occurring prior to purification. When HRG was treated with both plasmin and reducing agent, the protein was cleaved into several constituent parts as indicated by the band pattern on the gel, which included an intense band corresponding to the HRR/PRR fragment. This, together with the S-glutathionyl adduct observed at Cys185, provides direct evidence of redox regulation of HRG functioning in vivo.
Assessment of HRR/PRR release in serum-purified rabbit HRG. (A) SDS-PAGE analysis of rabbit HRG following treatment with or without plasmin and DTT. The HRR/PRR band is highlighted in the box and only appears under reducing conditions. (B) MS/MS analysis of the HRR/PRR band excised from the gel. Peptide sequences giving a confidence of 99% are in bold, and those with a 74% confidence in italics. The predicted sequence of the HRR/PRR plasmin cleavage product is highlighted in gray. NCBI accession number: XP_002716393. (C) Schematic showing release of HRR/PRR following both plasmin cleavage and reduction of the Cys185-Cys407 bond of HRG by GSH. The second interdomain disulfide bond between Cys6-Cys497 remains intact holding the remaining protein domains together.
Assessment of HRR/PRR release in serum-purified rabbit HRG. (A) SDS-PAGE analysis of rabbit HRG following treatment with or without plasmin and DTT. The HRR/PRR band is highlighted in the box and only appears under reducing conditions. (B) MS/MS analysis of the HRR/PRR band excised from the gel. Peptide sequences giving a confidence of 99% are in bold, and those with a 74% confidence in italics. The predicted sequence of the HRR/PRR plasmin cleavage product is highlighted in gray. NCBI accession number: XP_002716393. (C) Schematic showing release of HRR/PRR following both plasmin cleavage and reduction of the Cys185-Cys407 bond of HRG by GSH. The second interdomain disulfide bond between Cys6-Cys497 remains intact holding the remaining protein domains together.
Discussion
Due to the intrinsic disorder and heavy glycosylation, structural studies on HRG have been limited. The 1.93 Å x-ray crystal structure presented here is the first step toward structural characterization of HRG at the molecular level. In this study, crystallization was attempted using intact HRG purified from rabbit serum. Crystals of the N2 domain formed within the sample after 4 weeks. This relatively long crystallization period may be due to a need for the intact protein to slowly degrade releasing the N2 domain over this period of time. Although no proteases were added, flexible linker regions would be very susceptible to cleavage by even a small amount of any contaminating proteases that copurified from serum.
Both HRG N-domains (N1 and N2) are predicted to possess a cystatin-like fold based on their sequence homology to cystatin proteins.15 We show that indeed the N2 domain of rabbit HRG does have a cystatin-like structure that is composed of a 5-stranded antiparallel β-sheet, twisted around a 5-turn α-helix. The high degree of homology between rabbit and human HRG sequences (and conservation of Cys residues) suggests that our findings are also pertinent to the human protein. Although several structures of other cystatin proteins have been elucidated,33-35 the structure presented here is the first of a type 3 cystatin protein. Despite structural similarity to cystatins, HRG does not appear to exhibit protease inhibitory activity.15 The crystal structures of chicken egg white cystatin, pig cystatin A, and human cystatin B complexed with the protease papain, reveal the N-termini and 2 loops termed L1 (between β2 and β3) and L2 (between β3 and β4) to be important for their protease inhibition, with this segment forming a wedge shape that docks into the active site cleft of cysteine proteases.33-35 In HRG, the corresponding L2 loop in the N2 domain is extended by 6 amino acids, which we predict would preclude binding to papain-like proteases.
The N-terminal domains of HRG are also homologous with the heparin-binding region of antithrombin III.8,16 The helix of HRG could mimic the positively charged helical regions of the antithrombin heparin-binding site.36 Helical regions that bind to heparin are found in other proteins such as the protease nexin-1. The presented structure provides evidence that the N2 domains of HRG may contribute to the formation of HRG dimers in vivo but does not preclude the involvement of other domains in dimerization. It is important to acknowledge that the concentration of protein used for crystallization was higher than would normally be observed in plasma. Also in the full-length HRG protein, it is possible that other domains may disrupt the arrangement of the N2 domains in the crystal. If the N2 domain does in fact contribute to dimerization of the HRG molecule, it would significantly advance our understanding of the molecule. In this arrangement, the cysteine-GSH adducts are on the same face of the dimer that could allow interdomain disulfides to cross over between molecules; such an arrangement would permit cooperativity as reduction of 1 disulfide would destabilize both monomers within the dimer. The crystal structure of nexin-1 appears as 2 monomers and binds heparin between 2 helices, one from each monomer; these helices (corresponding to helix D in each monomer) interact with heparin through lysine residues.37 HRG regulates coagulation through neutralization of heparin and could mirror this binding mechanism, where the helices from the 2 cystatin-like domains, N1 and N2 (which also contain lysine residues) may together form a heparin-binding site. However, other domain combinations forming a heparin site (or sites) are possible, especially as the protein exists as a dimer under native conditions.
HRG is heavily glycosylated and the presented structure confirms Asn184 as an N-linked glycosylation site. SDS-PAGE analysis of crystals treated with deglycosylation enzymes confirmed that the N2 domain is glycosylated to a higher degree than observed in electron density maps; ∼13 kDa of its monomeric 72 kDa total mass corresponds to bound sugars (data not shown). Asn232 is annotated in the UniProt database (UniProt code: Q28640) as another potential glycosylation within this domain. However, there was no detectable glycosylation observed at this site in the structure. The observed S-glutathionyl adduct at Cys185 is direct evidence of an in vivo redox-regulated release mechanism for the antiangiogenic HRR/PRR fragment (Figure 4C). This implies that GSH reduces the Cys185-Cys407 disulfide bond in plasma such that the mixed disulfide occurs at Cys185 (and not at Cys407, which is present in the HRR/PRR fragment). It is known that plasma GSH concentrations, which are generally in the low micromolar range, are influenced by redox state.38 The HRG used in our experiments was purified from serum not plasma. To assess whether conversion of plasma to serum may have influenced redox activity at Cys185, we measured the free thiol concentration (which is indicative of redox state) in both rabbit serum and plasma. The free thiol concentration of the serum was found to be 208 µM, significantly lower than that of plasma, which was 250 µM (supplemental Figure 1A). HRG preparations derived from both plasma and serum were characterized and compared. Each preparation had a similar free thiol content (ca 0.7 mol/mol; supplemental Figure 1B) and molecular mass profile (supplemental Figure 2), indicating that the HRG proteins in each case are of the same molecular composition. These results confirm that the use of serum-derived HRG did not influence the redox state of the protein.
GSH-mediated posttranslational modification of proteins as a means of regulating angiogenesis is not without precedent. For example, S-glutathionylation of the low-molecular-weight protein tyrosine phosphatase negatively regulates angiogenesis through inhibition of vascular endothelial growth factor (VEGF)–regulated FAK activation.39 GSH is known to play other roles in regulating angiogenesis. It has been shown to prohibit ischemia-induced lung angiogenesis by sequestering of reactive oxygen species.40 GSH also regulates induction of hypoxia-inducible factor 1 (HIF-1), which controls the expression of proangiogenic genes such as VEGF.41 GSH was found to reduce HIF-1 binding activity and HIF-1–dependent promoter activity in squamous cell carcinoma cells in a dose-dependent manner.40
Plasmin-mediated cleavage of the HRG backbone is also required to release the antiangiogenic HRR/PRR fragment. The data here might suggest that generally reduction, and not proteolysis, occurs first given that sufficient quantities of reduced HRG were present to allow the crystal of the N2 domain to form, but this is not definitive. We did obtain evidence that a small proportion of the protein may undergo plasmin cleavage first by detecting the HRR/PRR fragment in serum-purified HRG treated with a reducing agent. This apparent cleavage may be due (at least in part) to the fact that the protein was extracted from serum given that a number of proteases, including plasmin, are activated during coagulation.42,43 As expected, the proportion of the HRR/PRR fragment was greatly increased by the addition of active plasmin to the sample. It has previously been demonstrated that HRG can aid activation of plasminogen to plasmin through formation of a complex with plasminogen and glycosoaminoglycans (GAGs) at the cell surface.44,45 Plasmin cleavage of human HRG has been shown to reduce its ability to bind cell-surface heparan sulfate.27 This has led to the idea that plasmin-mediated cleavage of HRG may act as a negative feedback mechanism that limits further plasminogen activation by reducing the amount of intact HRG that can efficiently tether plasminogen to cell-associated GAGs.27 The release of the antiangiogenic HRR/PRR fragment from HRG may therefore act to perturb proangiogenic effects associated with activated plasminogen.46 A gene knock-out study in mice has shown that plasminogen promotes vascular remodeling via both fibrinogen-dependent and -independent mechanisms.47 It is also plausible that by reducing the levels of intact HRG, the ability of the full-length protein to promote angiogenesis through its interaction with thrombospondin will be reduced. Therefore, it is likely that the breakdown of HRG and the release of the HRR/PRR fragment will negatively regulate angiogenesis by multiple mechanisms simultaneously.
In conclusion, the crystal structure presented provides a first molecular insight into HRG. It confirms that the N2 domain possesses a cystatin-like fold and is natively N-glycosylated at Asn184. Furthermore, the S-glutathionyl adduct at Cys185 provides direct evidence that the release of the antiangiogenic HRR/PRR fragment is partly controlled by a redox process, indicating a new additional role for GSH in controlling angiogenesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the British Heart Foundation, grant FS/10/036/28352 (A.J.S.). J.H.N. is a Senior Investigator of the Wellcome Trust (WT100209).
Authorship
Contribution: O.K., S.A.M., J.H.N., and A.J.S. conceived and designed the study; O.K., S.A.M., R.T., and C.H.B. collected data; O.K., S.A.M., R.T., C.H.B., J.H.N, and A.J.S. analyzed and interpreted the data; O.K., S.A.M., C.H.B., J.H.N, and A.J.S. drafted the manuscript; and all authors critically reviewed the manuscript and approved the final version.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alan J. Stewart, Medical and Biological Sciences Building, University of St. Andrews, North Haugh, St. Andrews, KY16 9TF, United Kingdom; e-mail: ajs21@st-andrews.ac.uk; and James H. Naismith, Biomedical Sciences Research Complex, University of St. Andrews, North Haugh, St. Andrews, KY16 9ST, United Kingdom; e-mail: jhn@st-andrews.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal