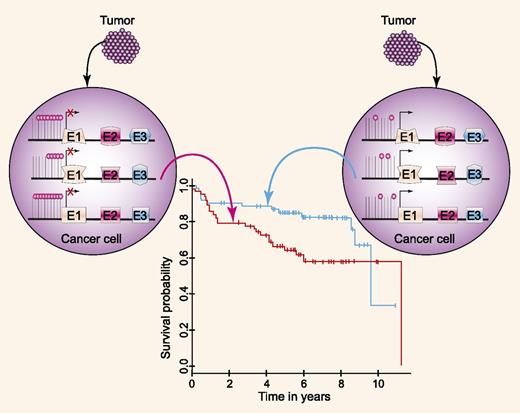
Magnitude of DNA methylation changes in DLBCL vs normal GC lymphocytes correlates with patients' survival. For source of the survival curve, see Figure 3B of Chambwe et al.1 Professional illustration by Paulette Dennis.
Magnitude of DNA methylation changes in DLBCL vs normal GC lymphocytes correlates with patients' survival. For source of the survival curve, see Figure 3B of Chambwe et al.1 Professional illustration by Paulette Dennis.
DLBCL, the most common subtype of non-Hodgkin lymphoma, is highly diverse from both biological and clinical standpoints. DLBCL pathogenesis is a complex multistep process that involves collaboration between the biological programs of normal B cells and acquired somatic tumor–associated genetic aberrations, including chromosomal translocations, gene amplifications, insertions/deletions and mutations, and posttranscriptional regulation by aberrantly expressed microRNAs.2,-4 Some of these genetic aberrations, as well as the expression of individual genes or gene signatures, may also modulate tumor aggressiveness and response to therapy, thus serving as biomarkers that predict or correlate with patients’ survival.5
Epigenetic changes, including epigenetic modifications of chromatin as well as aberrant hypermethylation or hypomethylation of promoters, may also contribute to lymphoma pathogenesis. Indeed, global DNA hypomethylation or focal changes in the methylation of promoters are observed in cancer. Previous methylome studies in DLBCL identified specific patterns of abnormal methylation, varying depending upon chromosomal regions, gene density, and the status of neighboring genes.6 Further, these studies identified distinctive epigenetic profiles in activated B-cell (ABC) and germinal center B-cell (GCB) DLBCL.7 However, these methylation patterns may reflect either the methylation state inherited from the normal cell of origin or methylation changes acquired during lymphomagenesis.
In this issue of Blood, Chambwe and colleagues carried out genomewide DNA methylation profiling of 140 DLBCLs and calculated the relative methylation difference between each case and normal GC lymphocytes.1 These studies revealed methylation variability between the genome of DLBCL and normal GC lymphocytes, identifying and quantifying the methylation changes likely acquired upon transformation. Further, the authors identified marked methylation variability across DLBCL tumors, and identified 6 clustering groups based on the magnitude of methylation changes. These DLBCL DNA methylation clusters were not exclusive of ABC vs GCB gene-expression subtype; however, each cluster was enriched in genes with common cellular functions. Moreover, for 5% to 14% of genes with perturbed methylation in each cluster, there was an inverse correlation with concordant RNA expression, indicating that the methylation changes affected expression of specific genes.
Previous studies have demonstrated that patients with GCB-like and ABC-like DLBCL tumors have different survivals that can be predicted by gene expression signatures.8 A correlation was also observed between patients’ survival and expression of specific genes within these signatures (eg, BCL6 and LMO2 for the GCB-like signature).9,10 To the best of our current knowledge, the work of Chambwe and colleagues is the first to show a correlation between the magnitude of DNA methylation changes and patient survival (see figure). Patients with a more disrupted DNA methylome compared with normal GC cells exhibited shorter survival than patients with fewer methylation changes. Interestingly, the pattern of methylation alone did not predict survival outcomes, suggesting that global and not specific methylation changes affected patient outcomes. The predictive value of the magnitude of DNA methylation changes and patients’ survival was independent of the International Prognostic Index. However, the authors did not evaluate its independence of the cell of origin classification. Although these findings are novel and imply that DNA methylation changes may underlie tumor aggressiveness and responsiveness to chemoimmunotherapy, these findings need to be validated in independent cohorts of patients, as is customary for predictive/prognostic biomarkers.5 This validation is important to determining the robustness and reproducibility of these findings and to eliminate the possibility of unintended model overfitting to the presently analyzed patient cohort.5 Even if validated in independent cohorts, the methodological complexity of the method employed by Chambwe et al will make it difficult to use this methylome biomarker in routine clinical practice. However, these findings will pave the way for studies looking at the impact of global methylation on tumor biology and responsiveness to therapy as a means of elucidating the mechanisms underlying changes in methylation magnitude in tumors. Addressing these questions will be the next important step in epigenetic studies that will further extend our knowledge of DLBCL pathogenesis.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal