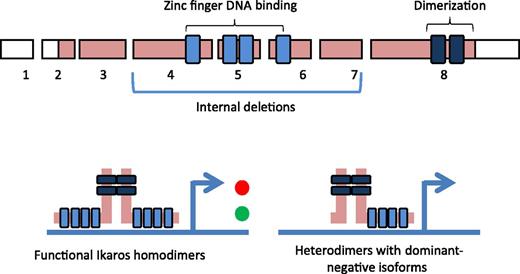
Structure of Ikaros. The N-terminal portion of Ikaros contains a DNA-binding domain with 4 zinc finger motifs. There are 2 zinc finger motifs at the C-terminal end that are essential for the formation of Ikaros homodimers. Ikaros binds directly to DNA to positively (green) or negatively (red) regulate expression of target genes. Internal deletions in exons 4 to 7 are common alterations that attenuate the DNA-binding capacity of Ikaros, and these deletions produce a dominant-negative isoform that results in loss of tumor-suppressor function of the WT allele, so that the transcriptional activity is lost (exons not drawn to actual scale).7
Structure of Ikaros. The N-terminal portion of Ikaros contains a DNA-binding domain with 4 zinc finger motifs. There are 2 zinc finger motifs at the C-terminal end that are essential for the formation of Ikaros homodimers. Ikaros binds directly to DNA to positively (green) or negatively (red) regulate expression of target genes. Internal deletions in exons 4 to 7 are common alterations that attenuate the DNA-binding capacity of Ikaros, and these deletions produce a dominant-negative isoform that results in loss of tumor-suppressor function of the WT allele, so that the transcriptional activity is lost (exons not drawn to actual scale).7
Treatment of children with BCR-ABL1–positive ALL has been revolutionized with the advent of TKI therapy. Historically, children with BCR-ABL1–positive ALL were treated with intensive chemotherapy followed by allogeneic hematopoietic stem cell transplantation (HSCT), and despite this, event-free survival rates of <50% were observed. With the advent of TKI therapy, outcomes have improved significantly,2,3 calling for a reassessment of the routine use of HSCT for BCR-ABL1–positive childhood ALL. With these emerging data, key questions remain about how to identify the highest-risk subsets of children with BCR-ABL1–positive ALL: how the use of TKI therapy might modulate this risk and how underlying pathogenic mechanisms portend a risk for treatment failure.
Deletions of IKZF1, a gene encoding the lymphoid transcription factor Ikaros, which is required for lymphoid development, are present in >70% of cases of BCR-ABL1–positive ALL.4 These alterations have been associated with poor outcomes both in children with BCR-ABL1–negative high-risk B-lineage ALL5 and in adults with BCR-ABL1–positive ALL,6 and this provided rationale for van der Veer et al to evaluate the prognostic significance of IKFZ1 deletions in pediatric patients with BCR-ABL1–positive ALL.1 Importantly, the authors examined 2 sequential cohorts of children, those treated both before and after the addition of imatinib to a common chemotherapeutic regimen and HSCT, to determine how TKI therapy might affect this risk.
Several key findings emerged from this study. The authors report that IKZF1 deletions have a negative prognostic impact in good-risk patients, where risk was defined by early treatment response, and this cannot be abrogated fully by the addition of imatinib. Although better than the 30% overall 4-year disease-free survival (DFS) rate for historically treated children without a TKI, the DFS for good-risk patients with IKZF1 deletions was only 55% on the EsPhALL protocol, which included imatinib. These findings highlight the need to further understand underlying pathogenic mechanisms in BCR-ABL1–positive ALL, because this subset of patients may benefit from additional targeted treatment. In contrast, good-risk, wild-type (WT) IKZF1 patients who received imatinib had outcomes comparable to those of children with newly diagnosed ALL without BCR-ABL1 with DFS rates of ∼75%, further supporting the deferral of HSCT for this group of pediatric patients.
Interestingly, IKZF1 deletions were not prognostic in poor-risk patients who received imatinib. DFS rates of ∼50% were observed, and there were no significant differences in DFS, overall survival, or relapse incidence between the WT and IKZF1-deleted poor-risk subsets. So although outcomes in patients with poor initial treatment responses did improve to some degree with TKI therapy, the presence of an IKZF1 deletion did not alter prognosis further, which again suggests that other underlying pathogenic factors are likely to be present in this group. Alternatively, perhaps more prolonged imatinib exposure could have improved outcomes for poor-risk patients.3
Although improvements in refining risk stratification are certainly warranted, the major thrust in current research is to define pathways for targeted therapy. In this regard, the key question is not whether but how IKZF1 deletions relate to poor outcomes in ALL. The inferior outcomes suggest that the gene product or affected pathways play some role in chemoresistance. The IKZF1 gene encodes for multiple isoforms, and the full-length product encodes DNA-binding zinc finger domains, a dimerization domain, and an activation domain (see figure).7 Ikaros proteins function to positively and negatively alter the transcription of downstream pathways in concert with activator or repressor complexes. IKZF1 deletions fall into 3 major categories: those that disrupt the protein coding exons completely (null mutations) on one allele, intragenic deletions that impact exons 4 to 7 that result in dominant-negative isoforms, and biallelic deletions that result in a null phenotype. Single base mutations also occur, many of which would be predicted to impair function.5
Although the contribution of each allele might vary, the functional impact of such alterations on the Ikaros pathway might be predicted to be (from most to less severe) biallelic/null, dominant negative, and haploinsufficient. In this scenario, the null and dominant-negative isoforms might be predicted to be associated with a worse prognosis, yet paradoxically, it was only haploinsufficient cases that conferred an adverse prognosis in the study by van der Veer et al. The authors speculate that this might be due to cases with large deletions, including monosomy 7, where the disruption of other pathways might contribute to the poor prognosis.
These intriguing observations need to be validated, because no such distinctions have been seen in previous studies6 and monosomy 7 deletions make up a minority of the IKZF1 disruptions. However, there is some evidence that the poorer prognosis of IKZF1 deletions might be related to the company it keeps. A recent report by this same group showed that additional copy-number alterations may confer a worse prognosis compared with cases with IKZF1 deletion alone.8 Finally, a very provocative report by Uckun et al found no evidence of diminished Ikaros protein expression or function in high-risk ALL, including BCR-ABL1–positive ALL.9 These authors suggest that the poor outcomes associated with IKFZ1 deletions in ALL could be a reflection of underlying genomic instability in aggressive leukemic clones rather than lost or diminished IKFZ1 function per se.
In summary, van der Veer et al have demonstrated that IKFZ1 deletions define a subset of BCR-ABL1–positive pediatric patients with unfavorable outcomes, despite treatment with contemporary TKI-based therapy, providing information that could potentially be used to alter treatment in the future. Their study also highlights the heterogeneity of this disease as well as the complexity of studies of IKFZ1 as a prognostic marker, because deletions have not been uniformly associated with poor outcomes in all subsets of patients.8,10 Questions still remain of whether outcome differences are directly related to loss of IKFZ1 gene function vs a reflection of other underlying pathogenic mechanisms. This study also provides further evidence for the good outcomes that can be achieved among favorable subsets of BCR-ABL1–positive patients with TKI therapy, supporting deferral of HSCT for this group.
Conflict-of-interest disclosure: The authors declare no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal