Key Points
KLF1 mutations cause severe congenital hemolytic anemia associated with a deficiency of red cell pyruvate kinase.
A severe KLF1 deficiency causes hereditary persistence of embryonic globin synthesis.
Abstract
In this study, we report on 8 compound heterozygotes for mutations in the key erythroid transcription factor Krüppel-like factor 1 in patients who presented with severe, transfusion-dependent hemolytic anemia. In most cases, the red cells were hypochromic and microcytic, consistent with abnormalities in hemoglobin synthesis. In addition, in many cases, the red cells resembled those seen in patients with membrane defects or enzymopathies, known as chronic nonspherocytic hemolytic anemia (CNSHA). Analysis of RNA and protein in primary erythroid cells from these individuals provided evidence of abnormal globin synthesis, with persistent expression of fetal hemoglobin and, most remarkably, expression of large quantities of embryonic globins in postnatal life. The red cell membranes were abnormal, most notably expressing reduced amounts of CD44 and, consequently, manifesting the rare In(Lu) blood group. Finally, all tested patients showed abnormally low levels of the red cell enzyme pyruvate kinase, a known cause of CNSHA. These patients define a new type of severe, transfusion-dependent CNSHA caused by mutations in a trans-acting factor (Krüppel-like factor 1) and reveal an important pathway regulating embryonic globin gene expression in adult humans.
Introduction
Anemia is a major public health problem affecting more than 1.5 billion people worldwide.1 The major causes of anemia include malnutrition, particularly iron and folate deficiency, and infections including malaria and HIV. However, a substantial proportion of patients with anemia have inherited red cell defects, with mutations found in genes encoding red cell enzymes, membrane proteins, heme, and globins. For example, every year more than 300 000 affected individuals are born with severe anemia resulting from inherited hemoglobinopathies.2 However, after screening for mutations in genes currently known to underlie red cell defects, there are many cases of inherited anemia in which either the patient’s clinical and hematological phenotype is not consistent with their described genotype or the cause of the anemia remains completely unknown.
Here we report on 8 unrelated patients who displayed severe, transfusion-dependent neonatal anemia with red cell abnormalities ranging from a thalassemia-like morphology (with hypochromic microcytic red cells) to that usually associated with chronic nonspherocytic hemolytic anemias (CNSHA) with anisopoikilocytosis, fragmented cells, and a high reticulocyte count. In addition, all of the probands had a remarkable persistence of fetal and embryonic globin synthesis. We found that the parents of these children also had significantly raised levels of fetal hemoglobin (HbF) consistent with previous observations in heterozygotes for KLF1 mutations. In the light of emerging evidence that mutations in KLF1, the gene encoding erythroid Krüppel-like factor (EKLF), give rise to persistent postnatal γ-globin expression in humans3-7 and embryonic globin expression in mice,8,9 we sequenced this gene in the probands. We discovered that all of them are compound heterozygotes for mutations in KLF1. Only 3 compound heterozygotes for KLF1 mutations have been previously reported, and their phenotypes have been only partially described.6,7 The transcription factor KLF1 is a master regulator of terminal erythroid differentiation, controlling expression of many key pathways and structures including cell division, the cell membrane and cytoskeleton, iron metabolism, and heme and globin synthesis.5 Here, we show that individuals with mutations affecting both alleles of KLF1 exhibit severe anemia, in which 1 or more of these pathways is significantly disrupted, giving rise to a wide spectrum of hematological phenotypes. The syndromes described here add to an increasing list of trans-acting mutations that cause unusual forms of inherited anemia.10-13 Most important, all such patients show persistent expression of substantial amounts of embryonic hemoglobins in adult life. Despite extensive characterization of the patterns of globin expression in millions of patients during the last 30 years, persistent expression of embryonic globins in adults has very rarely been reported.
Materials and methods
Blood samples
Blood was collected from all patients and parents after they signed a written informed consent. Patients receiving regular blood transfusion had a period of at least 10 weeks without blood transfusions before sample collection. This study was approved by the local ethical committees at Siriraj Hospital, Bangkok, Thailand, and at the John Radcliffe Hospital, Oxford, United Kingdom. This study also was conducted in accordance with the Declaration of Helsinki.
Hematology and blood serum factors
Routine hematology was determined using an automated red blood cell counter (Sysmex F280). Hemoglobin was analyzed by liquid chromatography, using an automated hemoglobin analyzer (HB Gold; Cumbria) and by isoelectric focusing (IEF) (Resolve; PerkinElmer). Abnormal hemoglobins were excised from the IEF gel, digested with trypsin and analyzed using a matrix-assisted laser desorption/ionization time-of flight (MALDI-TOF/TOF) mass spectrometer (Ultralex Bruker Daltonics). Routine biochemical parameters were measured using standard techniques.
Red cell phenotype analysis
Samples were tested for the Lutheran blood group using a gel card (DiaMed) with other additional blood group antigens including P1, Lea, Leb, k, Kpa, Kpb, Jka, Jkb, M, N, S, s, Fya, and Fyb, as per manufacturer’s instructions. The presence of Band-3 (AE-1 anion exchanger) on the red blood cell surface was estimated by the eosin-5-maleimide binding assay.14
Red blood cell enzyme activities for glucose-6-phosphate dehydrogenase, 6-phosphogluconate dehydrogenase, pyruvate kinase (PK), hexokinase, glucose phosphate isomerase, phosphofructokinase, aldolase, and phosphoglycerate kinase were assessed as described previously.15 Because reticulocytes contain higher amounts of these enzymes than mature red blood cells, a calculation for the net activities of each enzyme by correction for the reticulocyte number was performed to compare the results with our standard reference range.15
Flow cytometry to assess the proportions of HbF,16 Hb Bart’s (γ4), Hb ζ (ζ), and CD44 was performed on mature erythrocytes (CD71−) and circulating erythroblasts (CD71+) of patients and parents. In addition, leukocytes were stained with CD44 and CD45 after lysis of erythrocytes with fluorescence-activated cell sorter lysis solution (BD Biosciences). The antibodies used were anti-HbF directly conjugated to fluorescein isothiocyanate (FITC; Caltag), anti-Hb Bart’s (γ4), anti-Hb ζ (ζ), CD44 PE, CD45 Per CP, and CD71 PE-Cy5 (BD Biosciences), as well as isotype controls (mouse immunoglobulin G FITC and FITC-conjugated F[ab′]2 anti-mouse globulin; Dako).
Molecular characterization of the globin, KLF1 and PKLR genes
We performed multiplex Gap-PCR17 and amplification-refractory mutation system–PCR assays for detection of common α-thalassemia mutations.18 Sequence analysis was performed on 1.3 kb of the α2- and α1-, 1.81 kb of the ζ-, 2.2 kb of the ε-, and 2.85 kb of the β-globin genes and hypersensitive site 2 of the β-globin locus control region (630 bp), all promoters of the Gγ- (807 bp), Aγ- (802 bp), and β-globin (1044 bp) genes, using standard techniques. All primer sequences are summarized in supplemental Table 6, available on the Blood Web site. A multiplex ligation-dependent probe amplification assay was performed to exclude large rearrangements of the β-globin cluster.19 The KLF1 and PKLR genes were amplified and sequenced as previously described.20,21
Primary erythroid cell culture and expression analysis
Erythroid cells were obtained using a 2-phase culture system22 and harvested once they were double-positive for CD71 and glycophorin A. RNA was extracted, reverse transcribed to generate double-stranded cDNA (Superscript Double Stranded cDNA Synthesis Kit; Invitrogen), and labeled with dCTP-Cy3 as the test sample; 500 ng sonicated genomic DNA was labeled with dCTP-Cy5 as the input (Bioprime DNA Labeling System; Invitrogen). Both test and input samples were applied to a custom-designed Agilent expression array, as described.23 cDNA was analyzed by real-time PCR assays obtained from Applied Biosystem’s Assay-on-Demand resource. Expression was calculated relative to a reference gene (PABPC1) and normal Thai subjects (n = 6) as controls, using the Offal method.24
Results
Clinical presentation
All 8 patients reported here presented with neonatal jaundice requiring phototherapy and, in 5 cases, blood transfusion. Most patients were anemic at presentation with marked hepatosplenomegaly. By 1 year of age, all patients had received at least a single blood transfusion, and subsequently, all of them required regular transfusion during the first years of life. Three patients required splenectomy caused by hypersplenism. A summary of the clinical presentations and disease severity of the 8 probands is presented in supplemental Table 1 and associated footnotes.
Hematological evaluation
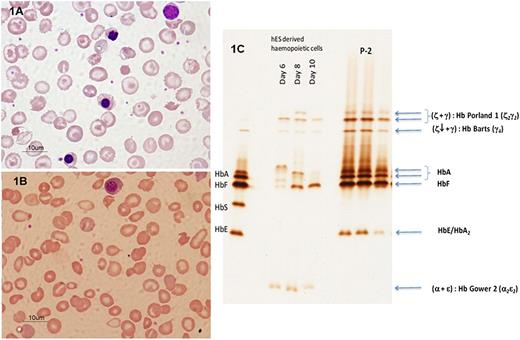
Full blood counts were determined for all 8 index patients and their parents and available siblings (Table 1). Despite transfusion, the levels of Hb in the probands (range, 3.0–8.3 g/dL) were low in some cases (Table 1). With a single exception (P1), all of the patients had hypochromic red cells (mean cell hemoglobin [MCH], 20.1–24.8 pg), suggesting abnormalities in the heme and/or globin synthesis pathways. All of the patients showed raised numbers of reticulocytes (5%–16%) and many nucleated red blood cells in the peripheral blood, suggesting a significant hemolytic component to the anemia. The morphology of the peripheral blood was variable. In some patients (P1, Figure 1A; P2–P4, supplemental Figure 1), the peripheral blood resembled that seen in patients with thalassemia, with marked hypochromasia, microcytosis, anisopoikilocytosis, target cells, and fragmented red cells. In those patients who coinherited globin gene defects, the severity of the changes was much greater than normally associated with these genotypes. In contrast, the peripheral blood of others (P5, Figure 1B; P6–P8, supplemental Figure 1) resembled that seen in nonspherocytic hemolytic anemia, with numerous fragmented red blood cells, schistocytes, and acanthocytes.
Hematological parameters, globin and KLF1 genotypes in Thai families with unusual forms of hereditary hemolytic anemia
| Cases . | Sex . | Age (y, mo) . | Hb (g/dL) . | Hct (%) . | RBC (×106/μL) . | MCV (fL) . | MCH (pg) . | MCHC (g/dL) . | RDW (%) . | Retic. (%) . | NRBC (/100WC) . | Hemoglobin typing (%) . | Globin genotypes . | KLF1 genotypes . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hb A . | Hb F . | HbA2/E . | Hb Portland-1 . | Hb Bart's . | αα/αα . | β/β . | Allele 1 . | Allele 2 . | ||||||||||||
| Family A | ||||||||||||||||||||
| P1 | F | 4* | 3.0 | 8.5 | 1.06 | 80.1 | 28.1 | 35.1 | 16.2 | 6.0 | 248 | 70.1 | 16.2 | 3.1 | ND | ND | αα/αα | β/β | R331W | G335R |
| 6.2† | 6.9 | 25.1 | 2.36 | 106.2 | 29.1 | 27.4 | 19.0 | 16.0 | 1510 | 38.5 | 51.8 | 2.0 | 4 | 2 | ||||||
| Father | M | 37 | 13.4 | 40.8 | 4.56 | 89.5 | 29.3 | 32.7 | 13.3 | 2.0 | 0 | 82.3 | 1.5 | 2.7 | 0 | 0 | αα/αα | β/β | W | G335R |
| Mother | F | 34 | 12.2 | 37.4 | 4.69 | 79.7 | 26.0 | 32.6 | 13.5 | 1.0 | 0 | 82.5 | 3.0 | 2.7 | 0 | 0 | αα/αα | β/β | R331W | W |
| Family B | ||||||||||||||||||||
| P2 | M | 0.6‡ | 5.9 | 18.2 | 2.76 | 65.9 | 21.4 | 32.4 | NA | 5.0 | 26 | 72.3¶ | 16.6 | 3 | 8.1§ | 0 | −SEA/αα | βE/β | G176RfsX179 | R301H |
| 12‖ | 7.5 | 25.7 | 3.72 | 69.1 | 20.1 | 29.1 | 17.8 | 9.0 | 181 | 16.9 | 39.1 | 4.3 | 16.2 | 14.7 | ||||||
| Father | M | 36 | 15.6 | 46.3 | 5.88 | 78.8 | 26.5 | 33.6 | 13.6 | 0.5 | 0 | 65.1 | 3.6 | 2.0 | 0 | 0 | αα/αα | β/β | W | R301H |
| Mother | F | 23** | 11.4 | 37 | 6.41 | 57.1 | 17.8 | 30.9 | ND | ND | 0 | 74.9 | 3.4 | 21.7 | 0 | 0 | −SEA/αα | βE/β | G176RfsX179 | W |
| Brother | M | 10 | 12.5 | 37.8 | 5.67 | 66.7 | 22.1 | 33.1 | 17.4 | 1.0 | 0 | 54.0 | 6.9 | 26.9 | 0 | 0 | αα/αα | βE/β | G176RfsX179 | W |
| Family C | ||||||||||||||||||||
| P3 | M | 0.2 | 5.2 | 19.0 | NA | 74.0 | 21.0 | 28.0 | 26.0 | 11.0 | 124 | ND | ND | ND | ND | ND | αα/αα | βE/βE | −154C/T | A298P |
| 1.10†† | 7.7 | 24.5 | 3.35 | 73.1 | 23.0 | 31.4 | 26.2 | 14.0 | 57 | 21.0 | 52.8 | 22.7 | + | + | ||||||
| Father | M | 31 | 14.0 | 42.1 | 5.45 | 77.3 | 25.7 | 33.3 | 14.5 | 1.0 | 0 | 68.4 | 0.5‡‡ | 31.1 | 0 | 0 | αα/αα | βE/β | W | A298P |
| Mother | F | 32 | 11.6 | 35.1 | 5.0 | 70.4 | 23.2 | 33.0 | 14.4 | 0.5 | 0 | 72.0 | 4.3‡‡ | 23.5 | 0 | 0 | αCSα/αα | βE/β | −154C/T | W |
| Family D | ||||||||||||||||||||
| P4 | M | 1.2¶¶ | 6.5 | 21.1 | 2.98 | 70.6 | 21.7 | 30.7 | 37.1 | 10.0 | 100 | 78.7 | 5.5 | 8.3 | 7.3 | 0.2 | −SEA/-α3.7 | βE/β | Q58X | A298P |
| 5.4§§ | 6.6 | 21.5 | 3.09 | 69.5 | 21.4 | 30.9 | 24.3 | 5.0 | 49 | 70 | 2.6 | 9.5 | 17.6 | 0.3 | ||||||
| Father | F | 28 | 14.0 | 42.5 | 6.98 | 60.9 | 20.1 | 32.9 | 17.2 | 1.5 | 0 | 75 | 2.6 | 22.4 | 0 | 0 | −SEA/αα | βE/β | Q58X | W |
| Mother | M | 27 | 12.1 | 34.6 | 4.81 | 72.0 | 25.2 | 35 | 15.5 | 1.0 | 0 | 70.2 | 1.8 | 28 | 0 | 0 | αα/-α3.7 | βE/β | W | A298P |
| Family E | ||||||||||||||||||||
| P5 | M | 18‖‖ | 3.4 | 11.1 | 1.37 | 81.0 | 24.8 | 30.6 | 25.4 | 10.1 | 113 | 62.7 | 29.0 | 4.6 | 10.2 | 6.0 | αα/αα | β/β | G176RfsX179 | A298P |
| 23*** | 8.2 | 27.4 | 3.38 | 81.1 | 24.2 | 29.9 | 22.4 | 6.0 | 86 | |||||||||||
| Father | M | 58 | 14.4 | 43.7 | 5.4 | 81.0 | 26.6 | 33.0 | 14.6 | 1.0 | 0 | 82.8 | 0.9 | 3.8 | 0 | 0 | −α3.7/αα | β/β | W | A298P |
| Mother | F | 55 | 14.1 | 42.7 | 4.85 | 88.0 | 29.1 | 33.1 | 13.9 | 1.0 | 0 | 82.8 | 3.1 | 2.8 | 0 | 0 | αα/αα | β/β | G176RfsX179 | W |
| Family F | ||||||||||||||||||||
| P6 | F | 0.9††† | 6.6 | 21.0 | 2.94 | 71.0 | 22.3 | 31.3 | 23.8 | 10.0 | 8 | 33.3 | 49.8 | 2.4 | 13.7 | 0.8 | αCSα/αα | β/β | G176RfsX179 | A298P |
| 5.7‡‡‡ | 8.3 | 27.1 | 3.82 | 70.9 | 21.6 | 30.5 | 23.4 | 10.0 | 13 | 45.0 | 50.0 | 3.0 | + | + | ||||||
| Father | M | 34 | 13.2 | 40.1 | 5.5 | 73.0 | 24.0 | 32.9 | 14.4 | 6.0 | 0 | 83.0 | 0.2 | 2.7 | 0 | 0 | αCSα/αα | β/β | W | A298P |
| Mother | F | 30 | 13.9 | 41.5 | 5.2 | 80.0 | 26.6 | 33.5 | 14.4 | 2.0 | 0 | 94.8 | 2.4 | 2.8 | 0 | 0 | αα/αα | β/β | G176RfsX179 | W |
| Brother 1 | M | 11 | 11.8 | 36.7 | 5.2 | 70.4 | 22.7 | 32.3 | 15.1 | 1.0 | 0 | 96.4 | 0.5 | 3.1 | 0 | 0 | αCSα/αα | β/β | G176RfsX179 | W |
| Brother 2 | M | 8 | 11.7 | 35.1 | 4.7 | 75.4 | 25.1 | 33.3 | 15.1 | 1.0 | 0 | 94.9 | 2.4 | 2.7 | 0 | 0 | αα/αα | β/β | W | A298P |
| Family G | ||||||||||||||||||||
| P7 | M | 0.2¶¶¶ | 5.7 | 17.8 | 2.66 | 67.0 | 21.2 | 31.9 | 21.9 | 11.0 | 146 | 33.9 | 54.6 | 2.2 | 14.3 | 0.3 | −α3.7/αα | β/β | G176RfsX179 | A298P |
| Mother | F | 22 | 11.9 | 36.4 | 4.59 | 79 | 26.0 | 32.8 | 14.4 | 1.0 | 0 | 85.8 | 0 | 2.6 | 0 | 0 | −α3.7/αα | β/β | G176RfsX179 | W |
| Family H | ||||||||||||||||||||
| P8 | M | 4§§§ | 5.4 | 16.3 | 2.18 | 75.0 | 24.7 | 33.1 | 20.4 | 10 | 26 | 69.0 | 19.0 | 10.0 | + | + | αα/αα | βE/β | G176RfsX179 | A298P |
| Father | M | 32 | 13.5 | 45.2 | 5.1 | 66.0 | 19.9 | 29.9 | 16.1 | 2.0 | 0 | 95.9 | 0.9 | 3.2 | 0 | 0 | −SEA/αα | β/β | W | A298P |
| Mother | F | 30 | 12.7 | 40.7 | 5.53 | 74.0 | 23.0 | 31.5 | 15.1 | 2.0 | 0 | 66.6 | 4.7 | 28.7 | 0 | 0 | αα/αα | βE/β | G176RfsX179 | W |
| Cases . | Sex . | Age (y, mo) . | Hb (g/dL) . | Hct (%) . | RBC (×106/μL) . | MCV (fL) . | MCH (pg) . | MCHC (g/dL) . | RDW (%) . | Retic. (%) . | NRBC (/100WC) . | Hemoglobin typing (%) . | Globin genotypes . | KLF1 genotypes . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hb A . | Hb F . | HbA2/E . | Hb Portland-1 . | Hb Bart's . | αα/αα . | β/β . | Allele 1 . | Allele 2 . | ||||||||||||
| Family A | ||||||||||||||||||||
| P1 | F | 4* | 3.0 | 8.5 | 1.06 | 80.1 | 28.1 | 35.1 | 16.2 | 6.0 | 248 | 70.1 | 16.2 | 3.1 | ND | ND | αα/αα | β/β | R331W | G335R |
| 6.2† | 6.9 | 25.1 | 2.36 | 106.2 | 29.1 | 27.4 | 19.0 | 16.0 | 1510 | 38.5 | 51.8 | 2.0 | 4 | 2 | ||||||
| Father | M | 37 | 13.4 | 40.8 | 4.56 | 89.5 | 29.3 | 32.7 | 13.3 | 2.0 | 0 | 82.3 | 1.5 | 2.7 | 0 | 0 | αα/αα | β/β | W | G335R |
| Mother | F | 34 | 12.2 | 37.4 | 4.69 | 79.7 | 26.0 | 32.6 | 13.5 | 1.0 | 0 | 82.5 | 3.0 | 2.7 | 0 | 0 | αα/αα | β/β | R331W | W |
| Family B | ||||||||||||||||||||
| P2 | M | 0.6‡ | 5.9 | 18.2 | 2.76 | 65.9 | 21.4 | 32.4 | NA | 5.0 | 26 | 72.3¶ | 16.6 | 3 | 8.1§ | 0 | −SEA/αα | βE/β | G176RfsX179 | R301H |
| 12‖ | 7.5 | 25.7 | 3.72 | 69.1 | 20.1 | 29.1 | 17.8 | 9.0 | 181 | 16.9 | 39.1 | 4.3 | 16.2 | 14.7 | ||||||
| Father | M | 36 | 15.6 | 46.3 | 5.88 | 78.8 | 26.5 | 33.6 | 13.6 | 0.5 | 0 | 65.1 | 3.6 | 2.0 | 0 | 0 | αα/αα | β/β | W | R301H |
| Mother | F | 23** | 11.4 | 37 | 6.41 | 57.1 | 17.8 | 30.9 | ND | ND | 0 | 74.9 | 3.4 | 21.7 | 0 | 0 | −SEA/αα | βE/β | G176RfsX179 | W |
| Brother | M | 10 | 12.5 | 37.8 | 5.67 | 66.7 | 22.1 | 33.1 | 17.4 | 1.0 | 0 | 54.0 | 6.9 | 26.9 | 0 | 0 | αα/αα | βE/β | G176RfsX179 | W |
| Family C | ||||||||||||||||||||
| P3 | M | 0.2 | 5.2 | 19.0 | NA | 74.0 | 21.0 | 28.0 | 26.0 | 11.0 | 124 | ND | ND | ND | ND | ND | αα/αα | βE/βE | −154C/T | A298P |
| 1.10†† | 7.7 | 24.5 | 3.35 | 73.1 | 23.0 | 31.4 | 26.2 | 14.0 | 57 | 21.0 | 52.8 | 22.7 | + | + | ||||||
| Father | M | 31 | 14.0 | 42.1 | 5.45 | 77.3 | 25.7 | 33.3 | 14.5 | 1.0 | 0 | 68.4 | 0.5‡‡ | 31.1 | 0 | 0 | αα/αα | βE/β | W | A298P |
| Mother | F | 32 | 11.6 | 35.1 | 5.0 | 70.4 | 23.2 | 33.0 | 14.4 | 0.5 | 0 | 72.0 | 4.3‡‡ | 23.5 | 0 | 0 | αCSα/αα | βE/β | −154C/T | W |
| Family D | ||||||||||||||||||||
| P4 | M | 1.2¶¶ | 6.5 | 21.1 | 2.98 | 70.6 | 21.7 | 30.7 | 37.1 | 10.0 | 100 | 78.7 | 5.5 | 8.3 | 7.3 | 0.2 | −SEA/-α3.7 | βE/β | Q58X | A298P |
| 5.4§§ | 6.6 | 21.5 | 3.09 | 69.5 | 21.4 | 30.9 | 24.3 | 5.0 | 49 | 70 | 2.6 | 9.5 | 17.6 | 0.3 | ||||||
| Father | F | 28 | 14.0 | 42.5 | 6.98 | 60.9 | 20.1 | 32.9 | 17.2 | 1.5 | 0 | 75 | 2.6 | 22.4 | 0 | 0 | −SEA/αα | βE/β | Q58X | W |
| Mother | M | 27 | 12.1 | 34.6 | 4.81 | 72.0 | 25.2 | 35 | 15.5 | 1.0 | 0 | 70.2 | 1.8 | 28 | 0 | 0 | αα/-α3.7 | βE/β | W | A298P |
| Family E | ||||||||||||||||||||
| P5 | M | 18‖‖ | 3.4 | 11.1 | 1.37 | 81.0 | 24.8 | 30.6 | 25.4 | 10.1 | 113 | 62.7 | 29.0 | 4.6 | 10.2 | 6.0 | αα/αα | β/β | G176RfsX179 | A298P |
| 23*** | 8.2 | 27.4 | 3.38 | 81.1 | 24.2 | 29.9 | 22.4 | 6.0 | 86 | |||||||||||
| Father | M | 58 | 14.4 | 43.7 | 5.4 | 81.0 | 26.6 | 33.0 | 14.6 | 1.0 | 0 | 82.8 | 0.9 | 3.8 | 0 | 0 | −α3.7/αα | β/β | W | A298P |
| Mother | F | 55 | 14.1 | 42.7 | 4.85 | 88.0 | 29.1 | 33.1 | 13.9 | 1.0 | 0 | 82.8 | 3.1 | 2.8 | 0 | 0 | αα/αα | β/β | G176RfsX179 | W |
| Family F | ||||||||||||||||||||
| P6 | F | 0.9††† | 6.6 | 21.0 | 2.94 | 71.0 | 22.3 | 31.3 | 23.8 | 10.0 | 8 | 33.3 | 49.8 | 2.4 | 13.7 | 0.8 | αCSα/αα | β/β | G176RfsX179 | A298P |
| 5.7‡‡‡ | 8.3 | 27.1 | 3.82 | 70.9 | 21.6 | 30.5 | 23.4 | 10.0 | 13 | 45.0 | 50.0 | 3.0 | + | + | ||||||
| Father | M | 34 | 13.2 | 40.1 | 5.5 | 73.0 | 24.0 | 32.9 | 14.4 | 6.0 | 0 | 83.0 | 0.2 | 2.7 | 0 | 0 | αCSα/αα | β/β | W | A298P |
| Mother | F | 30 | 13.9 | 41.5 | 5.2 | 80.0 | 26.6 | 33.5 | 14.4 | 2.0 | 0 | 94.8 | 2.4 | 2.8 | 0 | 0 | αα/αα | β/β | G176RfsX179 | W |
| Brother 1 | M | 11 | 11.8 | 36.7 | 5.2 | 70.4 | 22.7 | 32.3 | 15.1 | 1.0 | 0 | 96.4 | 0.5 | 3.1 | 0 | 0 | αCSα/αα | β/β | G176RfsX179 | W |
| Brother 2 | M | 8 | 11.7 | 35.1 | 4.7 | 75.4 | 25.1 | 33.3 | 15.1 | 1.0 | 0 | 94.9 | 2.4 | 2.7 | 0 | 0 | αα/αα | β/β | W | A298P |
| Family G | ||||||||||||||||||||
| P7 | M | 0.2¶¶¶ | 5.7 | 17.8 | 2.66 | 67.0 | 21.2 | 31.9 | 21.9 | 11.0 | 146 | 33.9 | 54.6 | 2.2 | 14.3 | 0.3 | −α3.7/αα | β/β | G176RfsX179 | A298P |
| Mother | F | 22 | 11.9 | 36.4 | 4.59 | 79 | 26.0 | 32.8 | 14.4 | 1.0 | 0 | 85.8 | 0 | 2.6 | 0 | 0 | −α3.7/αα | β/β | G176RfsX179 | W |
| Family H | ||||||||||||||||||||
| P8 | M | 4§§§ | 5.4 | 16.3 | 2.18 | 75.0 | 24.7 | 33.1 | 20.4 | 10 | 26 | 69.0 | 19.0 | 10.0 | + | + | αα/αα | βE/β | G176RfsX179 | A298P |
| Father | M | 32 | 13.5 | 45.2 | 5.1 | 66.0 | 19.9 | 29.9 | 16.1 | 2.0 | 0 | 95.9 | 0.9 | 3.2 | 0 | 0 | −SEA/αα | β/β | W | A298P |
| Mother | F | 30 | 12.7 | 40.7 | 5.53 | 74.0 | 23.0 | 31.5 | 15.1 | 2.0 | 0 | 66.6 | 4.7 | 28.7 | 0 | 0 | αα/αα | βE/β | G176RfsX179 | W |
Data of hemoglobin (Hb) analyses showed here derived from liquid chromatography in which HbA2 and Hb E were eluted at the same window and could not separate based on this methodology. Hb Portland-1 (ζ2γ2) and Hb Bart’s (γ4) have not been quantified in every patient (supplemental Table 1 and associated clinical text). Individuals without globin gene mutations are highlighted in gray.
F, female; M, male; MCH, mean cell hemoglobin; MCHC, mean cell hemoglobin concentration; MCV, mean corpuscular volume; NA, not available; ND, not determined; RDW, red cell distribution width; P, patient; Retic., reticulocyte count; W, wild type for KLF1 gene sequences.
This study was done when this patient was first referred to our hospital and 2 months after the last blood transfusion.
This study was performed 1 year after splenectomy and when the patient was free from blood transfusion. Of note, P1 is the only patient with a normal MCH (28 pg), suggesting that the coinheritance of thalassemia makes a significant contribution to the defects in Hb synthesis seen in our patients.
This study was performed when the patient (P2) was first seen at our hospital in 1997 at age 6 months.
The patient received less than 1 month of blood transfusion before this study and has been on regular transfusion since.
At first diagnosis, this abnormal hemoglobin was identified as Hb Bart’s.
This study was performed 6 months after splenectomy.
The mother of P2 died of lung cancer on February 18, 2004.
The patient was withheld from regular blood transfusion for 4 months before this study.
HbF measurements were variable even within the same individual; at 2 years before this study, the Hb F levels in the mother and the father were 1.5% and 1.2%, respectively by low-pressure liquid chromatography (LPLC; same laboratory). Moreover, the Hb F levels measured were different based on the techniques used; in the mother, Hb F levels were 4.3%, 3.4%, and 3.3%, and those of the father were 0.5%, 0.9%, and 0%, based on LPLC, high-performance liquid chromatography, and capillary electrophoresis, respectively. Using capillary electrophoresis, we were able to discriminate between Hb E and HbA2; interestingly, the levels of HbA2 in the mother were 3.8% (with 20.8% Hb E) compared with 5.4% (with 26.1% Hb E) in the father. In addition, 0.2% of Hb Constant Spring eluted at retention time 305 s was identified in the mother.
This study was done 2 months after blood transfusion, when this patient was first referred.
This study was performed 3 months after the regular blood transfusion was held.
This study was performed when this patient was withheld from blood transfusion for 10 weeks.
This study was performed 6 months after splenectomy.
This study was performed when this patient first presented at our hospital.
This study was performed after the patient was free from blood transfusion for 2 years.
This study was performed when this patient first presented at age 2 months.
This evaluation was done when the patient was first referred to our center, and the last blood transfusion was performed around 12 weeks before this study.
Wright stained peripheral blood pictures from 2 index patients who had normal α- and β-globin genes. In P1 (A), a marked hypochromic microcytosis with anisopoikilocytosis, acanthocytes, and numerous nucleated red blood cells was observed; in P5 (B), a blood picture with numerous fragmented red blood cells with schistocytes and numerous nucleated red blood cells was seen, similar to nonspherocytic hemolytic anemia. Both smears were performed after splenectomy in both patients and were free from blood transfusion. Peripheral blood features in other patients are available in supplemental Figure 1. (C) IEF study of embryonic globins identified in a patient with KLF1 mutations. Comparing hemoglobin profiles from the patient, P2, with control human embryonic stem cell (hES)-derived hematopoietic cells reveals 3 distinct abnormal hemoglobin protein bands separated at a more cathodic position than HbA and HbF. These hemoglobin species were similar to those of patients (P2, in triplication), as they were separated to the same isoelectric points. These hemoglobin bands were subsequently identified by mass spectrometry to be Hb Portland-1 (ζ2γ2) and Hb Bart’s (γ4). Of note, a different level of embryonic protein expression during erythroid differentiation from embryonic to fetal erythropoiesis in hES cells from day 6 to day 10 was observed (Right). Moreover, a fast-moving hemoglobin specie of Hb Gower 2 (α2ε2) was identified in this erythroid cell model but not from the patient. The standard hemoglobin controls are shown on the far left lane.
Wright stained peripheral blood pictures from 2 index patients who had normal α- and β-globin genes. In P1 (A), a marked hypochromic microcytosis with anisopoikilocytosis, acanthocytes, and numerous nucleated red blood cells was observed; in P5 (B), a blood picture with numerous fragmented red blood cells with schistocytes and numerous nucleated red blood cells was seen, similar to nonspherocytic hemolytic anemia. Both smears were performed after splenectomy in both patients and were free from blood transfusion. Peripheral blood features in other patients are available in supplemental Figure 1. (C) IEF study of embryonic globins identified in a patient with KLF1 mutations. Comparing hemoglobin profiles from the patient, P2, with control human embryonic stem cell (hES)-derived hematopoietic cells reveals 3 distinct abnormal hemoglobin protein bands separated at a more cathodic position than HbA and HbF. These hemoglobin species were similar to those of patients (P2, in triplication), as they were separated to the same isoelectric points. These hemoglobin bands were subsequently identified by mass spectrometry to be Hb Portland-1 (ζ2γ2) and Hb Bart’s (γ4). Of note, a different level of embryonic protein expression during erythroid differentiation from embryonic to fetal erythropoiesis in hES cells from day 6 to day 10 was observed (Right). Moreover, a fast-moving hemoglobin specie of Hb Gower 2 (α2ε2) was identified in this erythroid cell model but not from the patient. The standard hemoglobin controls are shown on the far left lane.
The parents and siblings of the 8 affected individuals were not anemic (Hb, 11.4–15.6 g/dL), although many of them had abnormal red cell indices because they are carriers for common forms of thalassemia in Thailand (see following). In the 6 individuals (highlighted in gray in Table 1) who were not carriers of hemoglobinopathies (αα/αα:β/β), the red cell indices were either unremarkable or marginally hypochromic and microcytic (average MCH, 27.1 + 1.71 pg [range, 25.1–29.3 pg]; average mean corpuscular volume, 81.9 + 5.57 fL [range, 75.4–89.5 fL]).
Analysis of hemoglobin
Hemoglobin analysis revealed abnormally high levels of HbF (average, 38%; range, 2.6%–54.6%) in all patients, consistent with increased F cells by flow cytometry (heterocellular pattern; supplemental Table 2). Two unanticipated hemoglobins present at levels of up to 18% were observed in all patients. Using IEF and MALDI-TOF mass spectrometry (Figure 1B, supplemental Figure 2, and Table 1), these hemoglobins were identified as the γ chain tetramer Hb Bart’s (γ4) and the embryonic hemoglobin Portland I (ζ2γ2). The presence of both Hbs was further confirmed by flow cytometry (supplementary Table 2). Hb Gower 2 (α2ε2) was not detected at significant amounts, and other minor bands (which may correspond to Hb Gower 1, ζ2ε2) were not examined further. Increased levels of embryonic ζ and ε-globin mRNA expression were observed by analysis of cDNA extracted from the peripheral blood of patient P2, using a tiled microarray (Figure 2A-B), as well as in other 5 patients using real-time PCR assays (Figure 2C). Interestingly, significantly increased proportions of HbF were observed in 14 of 15 parents from the index families (range, 0.2%–4.7%; average (SD), 2.34% + 1.34%; Table 1), but none of the parents expressed Hb Bart’s or embryonic globins, as assessed by chromatography and IEF (Table 1). Detection of F-cells, anti-ζ, and anti-Bart’s using flow cytometry was variable in these parents and siblings (supplemental Table 2).
Expression profiles of the primary erythroid culture cells from patients with KLF1 mutations. An expression profiling of the α(A) and β(B) globin gene clusters using Agilent tiled oligonucleotide array was performed in P2 erythroid cells compared with normal control. (A) A distinctive higher peak of ζ globin gene expression (dark arrow with gray box highlighted) was observed in the patient (bottom) compared with a normal cDNA from a control sample (top). (B) Two unique upregulated expression peaks of γ-globin (arrow 1) and ε-globin (arrow 2) genes in the patient were shown. Another upregulating expression of noncoding mRNA (arrow 3) was observed at the β-globin locus control region (β-globin locus control region, hypersensitive site 2 [HS-2]), all with dark arrows and gray boxes highlighted. However, the meaning of this observation on the downstream globin gene regulation and hereditary persistence of embryonic globin in the patient remains unclear. There was no significant change on expression of other nearby genes on both clusters. (C) Expression profile of erythroid specific genes from primary erythroid cell samples from 5 patients (P) with KLF1 mutations and normal controls (n = 6), using qPCR and Taqman probe hybridization, showing a marked increase of fetal and embryonic globin mRNA expression (HBZ, ζ-globin gene; HBE, ε-globin gene; HBG, γ-globin gene) in all patients with relatively normal expression of KLF1. This suggests that although most patients carry a single nonsense mutation, which could result in haplo-insufficiency, a missense mutation on the other allele might produce stable transcripts that could be detected at similar levels to normal. Alternatively, our mRNA analysis used might not be sensitive enough to demonstrate a minor perturbation in the level of KLF1 expression. With the exception of BCL11A (B-cell CLL/lymphoma 11A [zinc finger protein] from 1 patient [P2]), there was no change in expression compared with normal controls of other erythroid-specific genes, including CD71, SOX6, ERAF, GYPA, glycophorin A, and EPOR (erythropoietin receptor; supplementary Figure 5).
Expression profiles of the primary erythroid culture cells from patients with KLF1 mutations. An expression profiling of the α(A) and β(B) globin gene clusters using Agilent tiled oligonucleotide array was performed in P2 erythroid cells compared with normal control. (A) A distinctive higher peak of ζ globin gene expression (dark arrow with gray box highlighted) was observed in the patient (bottom) compared with a normal cDNA from a control sample (top). (B) Two unique upregulated expression peaks of γ-globin (arrow 1) and ε-globin (arrow 2) genes in the patient were shown. Another upregulating expression of noncoding mRNA (arrow 3) was observed at the β-globin locus control region (β-globin locus control region, hypersensitive site 2 [HS-2]), all with dark arrows and gray boxes highlighted. However, the meaning of this observation on the downstream globin gene regulation and hereditary persistence of embryonic globin in the patient remains unclear. There was no significant change on expression of other nearby genes on both clusters. (C) Expression profile of erythroid specific genes from primary erythroid cell samples from 5 patients (P) with KLF1 mutations and normal controls (n = 6), using qPCR and Taqman probe hybridization, showing a marked increase of fetal and embryonic globin mRNA expression (HBZ, ζ-globin gene; HBE, ε-globin gene; HBG, γ-globin gene) in all patients with relatively normal expression of KLF1. This suggests that although most patients carry a single nonsense mutation, which could result in haplo-insufficiency, a missense mutation on the other allele might produce stable transcripts that could be detected at similar levels to normal. Alternatively, our mRNA analysis used might not be sensitive enough to demonstrate a minor perturbation in the level of KLF1 expression. With the exception of BCL11A (B-cell CLL/lymphoma 11A [zinc finger protein] from 1 patient [P2]), there was no change in expression compared with normal controls of other erythroid-specific genes, including CD71, SOX6, ERAF, GYPA, glycophorin A, and EPOR (erythropoietin receptor; supplementary Figure 5).
Analysis of structural proteins commonly mutated in red cell disorders
Only 2 of the probands (P1 and P5) were found to have entirely structurally normal α- and β-globin genes. The other patients were single or double heterozygotes for common mutations of the globin genes. However, their clinical severity far exceeded that seen in their relatives or other carriers of thalassemia. All patients, and many of the parents, had unexplained increased levels of HbF, which could have been linked to a deletion or deletions involving the β-globin locus.25 However, we excluded the possibility of mutations lying within the cis-regulatory regions involved in γ-globin gene expression using multiplex-ligation probe assays and sequencing analysis (supplementary Figure 3 and supplementary Table 3).
Although the hematologic profiles in these patients were complex, 4 patients (P5–P8) were initially diagnosed with red cell enzymopathies.26 These patients were tested for 8 common red cell enzyme activities and were found to have significant PK deficiency (<50% of normal after reticulocyte count correction). On the basis of these findings, we evaluated the PK status of the remaining 4 patients (P1–P4), in whom we did not initially suspect any enzymopathy. All had significantly reduced levels of PK activity (supplementary Table 2). However, DNA sequencing of the coding region including the intron/exon boundaries of the PKLR gene, encoding the red cell PK enzyme, failed to identify any causative mutations.
We next evaluated expression of proteins present on the red cell membrane and in the cytoskeleton. Extensive minor blood group analysis in 5 index families is summarized in supplementary Table 4. All patients had the rare In(Lu) blood group phenotype (a−/b−). In addition, all parents and siblings, except one (the father of P2), also expressed this rare blood group phenotype. The In(Lu) phenotype results from the suppression of 2 cell surface glycoproteins, BCAM and CD44, which carry the Lutheran and Indian blood group antigens, respectively.21,27 It has previously been shown that carriers of KLF1 mutations have the In(Lu) phenotype and that expression of CD44 was significantly reduced in the erythrocytes of the majority of patients tested (supplementary Table 2), but not in their leukocytes, whereas expression of the integral protein Band 3 evaluated by eosin-5-maleimide binding assay appeared unchanged compared with control samples (supplementary Table 2).
Identification of mutations in the KLF1 genes
Although the levels of embryonic (ζ and ε) and fetal (γ) globin RNA and protein are significantly elevated in all probands, analysis of their globin gene haplotypes showed that they have inherited quite different combinations of α- and β-globin haplotypes (supplemental Figure 6 and supplemental Figure 7). Given that elevations in embryonic globins resulting from cis-acting mutations have never been described previously, it seemed very unlikely that these patients had inherited a variety of cis-mutations causing persistent embryonic gene expression. To analyze this in further detail, the embryonic ζ and ε genes and the fetal γ genes were sequenced in all patients, but no changes (other than common single nucleotide polymorphisms) were found.
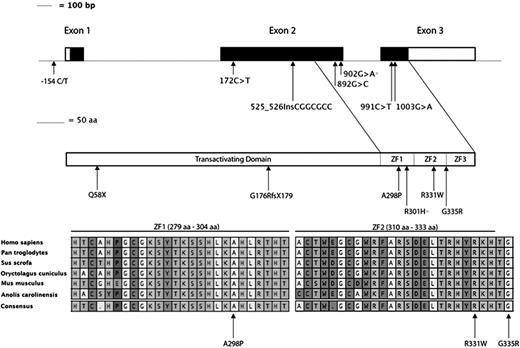
These observations led us to hypothesize that the genetic lesion in these patients may lie in a trans-acting factor involved in erythroid differentiation and maturation. Therefore, we DNA sequenced the coding region and intron/exon boundaries of 2 key erythroid transcription factors, GATA1 and KLF1, in a single proband (P2). No likely pathogenic changes were identified in the GATA1 gene; however, we found 2 changes in KLF1, a c.902G>A transition leading to a p.R301H substitution and a 7 bp insertion (c.525_526InsCGGCGCC), resulting in a frameshift (p.G176RfsX179) and disruption of the protein before the zinc finger DNA binding domain (Figure 3 and Table 1). Each of these sequence changes has a separate parental origin. The c.902G>A change has been previously reported to underlie elevated levels of HbF, as has a similar 7bp insertion (leading to p.G176AfsX179).6 To gain further genetic evidence that compound heterozygosity for deleterious KLF1 alleles may underlie the abnormalities reported here, we DNA sequenced this gene in the 7 remaining patients. Surprisingly, we found all 7 to be compound heterozygotes for likely pathogenic KLF1 changes (Figure 3, Table 1, and supplemental Figure 4). Four previously unreported coding changes were identified: a c.172C>T transition causing a premature truncation at position 58 in the protein (p.Q58X) before the zinc finger domains, a c.892G>C transversion predicted to alter alanine in the first zinc finger (p.A298P), a c.991C>T transition leading to a p.R331W amino acid change in the second zinc finger, and a c.1003G>A change leading to a p.G335R substitution that alters a glycine residue just outside the second zinc finger domain. All 3 novel missense changes identified alter residues that are highly conserved throughout evolution (Figure 3). We also identified a C>T change 154 bp 5′ of the initiating ATG codon (−154 C>T) in a single patient (P3). This region has previously been reported to include the promoter region of KLF120 , and the change is predicted to alter the binding of transcription factors tumor suppressor p53, paired box 5, and epidermal growth factor receptor–specific transcription factor (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3). This change was absent in 100 population-matched control individuals and is also absent from single nucleotide polymorphism database (http://www.ncbi.nlm.nih.gov/SNP/), suggesting it is rare. These genetic data, taken together with the patient’s phenotypic abnormalities, strongly suggest this change disrupts expression of the associated KLF1 transcript, as seen in a single patient (P1) tested (Figure 2C).
KLF1 gene structure, identification of variants in cases reported in this study, and their location within conserved domains. (top) Schematic representation of KLF1; exons are shown to scale, with the coding sequence in black and untranslated regions in white, and the locations of nucleotide changes identified here. Below this is a diagram representing the KLF1 protein with the previously annotated transactivating domain and the 3 zinc finger domains encoded by exons 2 and 3. The locations of all mutations identified in this study are shown; all changes are novel except for the previously reported R301H substitution (asterisk). (bottom) A position and conservation of amino acid residues found to be changed in the probands reported here (arrows). The entire first and second zinc finger sequences are shown with interspecies conservation, and the extent of each zinc finger domain is shown above each region. The cysteine and histidine residues involved in the coordination of zinc are denoted by the # symbol, and the residues that contact specific bases in DNA are marked with a + symbol.
KLF1 gene structure, identification of variants in cases reported in this study, and their location within conserved domains. (top) Schematic representation of KLF1; exons are shown to scale, with the coding sequence in black and untranslated regions in white, and the locations of nucleotide changes identified here. Below this is a diagram representing the KLF1 protein with the previously annotated transactivating domain and the 3 zinc finger domains encoded by exons 2 and 3. The locations of all mutations identified in this study are shown; all changes are novel except for the previously reported R301H substitution (asterisk). (bottom) A position and conservation of amino acid residues found to be changed in the probands reported here (arrows). The entire first and second zinc finger sequences are shown with interspecies conservation, and the extent of each zinc finger domain is shown above each region. The cysteine and histidine residues involved in the coordination of zinc are denoted by the # symbol, and the residues that contact specific bases in DNA are marked with a + symbol.
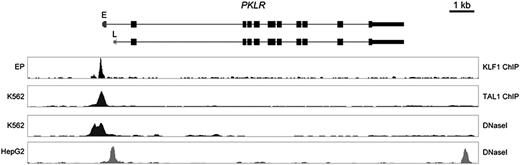
All parents and some siblings studied were heterozygotes for the KLF1 changes present in probands. The 4 coding changes are extremely rare and are listed neither in dbSNP136 (http://www.ncbi.nlm.nih.gov/projects/SNP/) nor in ∼13 000 alleles from African- and European-Americans listed in the Exome Variant Server (http://evs.gs.washington.edu/EVS/). In addition, both of these KLF1 changes were absent from 296 normal chromosomes from population-matched control individuals. The p.Q58X change is predicted to prematurely truncate the KLF1 protein, and the 3 novel missense changes alter invariant residues in the first and second zinc finger regions (Figure 3) and may disrupt DNA binding. Altered binding of KLF1 in vivo may underlie the observed PKLR deficiency in these patients as the WT KLF1 protein binds the promoter region of the PKLR gene in human erythroid progenitor cells (Figure 4).
Binding of KLF1 to the PKLR gene. (top) a representation of the PKLR gene structure including the start of the erythroid (E)-specific transcript (dark gray) and the liver (L)-specific first exon in light gray. (bottom) KLF1 binding and DNase I hypersensitive sites in cultured human erythroid progenitors (EP), K562; erythroleukemia cell line, HepG2; and liver cells line, as indicated.
Binding of KLF1 to the PKLR gene. (top) a representation of the PKLR gene structure including the start of the erythroid (E)-specific transcript (dark gray) and the liver (L)-specific first exon in light gray. (bottom) KLF1 binding and DNase I hypersensitive sites in cultured human erythroid progenitors (EP), K562; erythroleukemia cell line, HepG2; and liver cells line, as indicated.
KLF1 target genes
The predominant hematologic phenotypes in these compound heterozygotes appear to arise from the effects of KLF1 on the globin genes (causing abnormal globin synthesis) and the genes encoding CD44 (causing the In(Lu) phenotype) and PK (causing CNSHA). Of interest, on the basis of reanalysis of previously published data (supplemental Table 5), these erythroid genes are among the most sensitive to changes in the levels of KLF1. Others have previously analyzed the binding of KLF1 to cis-elements surrounding the globin genes and the CD44 gene.28 Here we have analyzed binding of KLF1 to the PKLR gene (based on data from Su et al29 showing that it specifically binds the erythroid promoter of this gene; Figure 4). This is clearly consistent with mutations in the KLF1 gene causing severe downregulation of the levels of PK resulting in CNSHA. It was of interest that all patients with the CNSHA phenotype and PK deficiency (P5–P8) have exactly the same combination of mutations (p.G176RfsX179;p.A298P; Table 1). Finally, it was of interest that in a single case (P2) that could be analyzed, the level of BCL11A mRNA was reduced (supplementary Figure 5) when both alleles of the KLF1 gene are mutated. This is consistent with previous observations showing that this gene is a direct target of KLF1 and is normally involved in the silencing of γ-globin synthesis.3,4 Following on from observations of the effect of BCL11A deficiency on embryonic globin expression in the mouse30 and the anecdotal observation that embryonic expression is increased in a patient with congenital dyserythropoietic anemia type IV associated with a dominantly acting KLF1 mutation (E325K),30 it seems plausible that when KLF1 is downregulated to a critical level, it also has a similar effect on human embryonic globin expression.
Discussion
We have defined a new cause of severe anemia in pediatric patients who are compound heterozygotes for mutations in the key erythroid transcription factor KLF1. The cardinal features of this condition are transfusion-dependent anemia associated with abnormalities in red cell enzymes (eg, PK deficiency), red cell membranes [In(Lu) phenotype], and an abnormal pattern of globin synthesis with very high levels of HbF and readily detectable levels of embryonic globins in adult life.
It appears that KLF1 mutations may cause a wide spectrum of phenotypes, which may have been expected from its pleiotropic role in erythropoiesis. The principles emerging from current studies suggest that both the levels of KLF1 and the type of mutated protein produced may exert different effects on KLF1 target genes and, consequently, cause a wide range of red cell phenotypes. The nonsense mutation G176RfsX179 was identified in 5 of the families reported here and is similar to the G176AfsX179 reported in 2 individuals from Korea and Vietnam,6 but no patients have inherited 2 nonsense mutations. It seems likely that this would lead to embryonic or early fetal lethality resulting from severe in utero anemia, as occurs in null mice.32 This warrants further studies of KLF1 in families with unexplained, recurrent hydrops fetalis or fetal loss, which cannot be accounted for by other conditions.
It has previously been shown that a mutation (E325K) in the second zinc finger domain of KLF1 causes a significant alteration in DNA binding, especially at the β-globin genes, resulting in a severe red cell and clinical phenotype (congenital dyserythropoietic anemia), even when only a single allele is mutated.30 It is thought that this mutation acts as a dominant negative, gain-of-function mutation. In contrast, the patients described here have newly defined phenotypes (severe thalassemia or CNSHA) associated with KLF1 mutations. It is of interest that all patients with CNSHA had exactly the same genotype (Table 1, Family E-H), suggesting that the phenotype is largely driven by the KLF1 genotype, rather than by epistatic effects. Of particular interest is that the A298P mutation lies in a key residue of the first zinc finger of KLF1, which determines the binding specificity of the protein.33
Given the widespread effects of KLF1 in erythropoiesis summarized here, it seems unlikely that this study and that of Satta et al7 have captured the full spectrum of these inherited anemias. Despite the apparent relationship between genotype and phenotype, it is interesting that all 3 previously described compound heterozygotes for KLF1 mutations (p.S270X/p.K332Q)7 and (G176Afs179X;L51R)6 had similarly raised levels of HbF (22%, 31%, and 9.5%), but rather milder anemia (Hb, 12.1, 11.5, and 13.7 g/dL), than the patients described here. However, 2 of these patients were adults (older than 30 years)7 when analyzed and also harbored a mutation (p.K322Q) that, in vitro, had only a mildly decreased DNA binding affinity to the promoters tested (∼2 fold), and therefore may represent a relatively mild hypomorphic allele.34 The age of the third patient is not reported. We have noticed (anecdotally) that anemia in our patients has improved with age, requiring progressively less transfusion. It will be important to follow-up all patients with KLF1 mutations to determine whether severity may be truly age-dependent.
During the last few years, the role played by KLF1 in globin gene regulation has become clearer.5,35 Almost since its discovery, it has been known that KLF1 is an activator of β-globin expression, but more recently, it also has been shown to play a role in silencing γ-globin expression, probably by regulating BCL11A and its interaction with Sox6 at the γ-globin gene.3,36,37 Mutations in KLF1 therefore reduce β-globin expression (causing β thalassemia)32 and de-repress γ-globin expression, thereby increasing the synthesis of HbF.4 However, the relationship between these effects and the mutations that downregulate KLF1 are not straightforward: Not all heterozygotes for KLF1 mutations produce increased levels of HbF,38 and in those who do, the level can be very different, even in the presence of the same mutation, demonstrating variable penetrance.7,37,38 These observations have led to speculation about the role of KLF in regulating the levels of HbF.36 The data presented here clarify the situation because all report that compound heterozygotes produce substantial amounts of HbF, putting beyond doubt that KLF1 plays a major role in normally repressing γ-globin synthesis in humans in vivo. Reactivation of γ-globin synthesis by downregulating KLF1 might provide a way to ameliorate the effect of severe β-hemoglobinopathies.3,39
In mice, which do not have a fetal stage of erythropoiesis, the KLF1/BCL11A circuit affects embryonic rather than fetal gene expression.40 This pathway normally suppresses embryonic globin synthesis in definitive erythroid cells, but a knockout of Klf1 appears to have no effect on embryonic globins in primitive erythropoiesis, when they are normally expressed. In this model, definitive cells are not produced to determine whether or not the embryonic genes are correctly suppressed later in development.5 It was reported anecdotally that a patient with congenital dyserythropoietic anemia type 4 and the E325K mutation had raised levels of embryonic globins. Here we show that in compound KLF1 heterozygotes, human embryonic (ζ and ε) globin expression persists in adults. This indicates that KLF1 also plays a role in repressing embryonic globin expression in humans. This finding is striking because reactivation of embryonic globin is virtually never seen in other erythroid disorders. At present, the pathway underlying embryonic globin repression is poorly understood. However, it has recently been shown that 2 members of the KLF family (KLF3 and KLF8) may be involved in regulating embryonic globin expression, and a knockout of these 2 transcription factors in erythroid cells de-represses embryonic globin synthesis.41 Because expression of these 2 factors is driven by KLF1, it is possible that mutations in this protein might reduce KLF3 and KLF8, leading to derepression of embryonic globins. Further analysis of this pathway is warranted because preliminary studies in mice suggest that persistence of embryonic globin expression could ameliorate severe hemoglobinothathies.42
In conclusion, we have shown that a wide spectrum of unexplained forms of severe inherited anemia may be caused by trans-acting mutations in the key erythroid transcription factor KLF1. Most remarkably, in every case where the downregulation is sufficiently severe to cause anemia, there is an associated persistence of embryonic globin expression revealing a new pathway by which globin gene expression might be manipulated to ameliorate other inherited hemoglobinopathies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank all patients and parents for their participation in this study and their permission to report our findings. We also thank Helena Ayyub, Douglas Vernimmen, Ram Malladi, Katalin di Gleria, Jennifer Eglinton, and Portep Tanyut for technical support and Professor Wachara Kasinlerk of Chiang Mai University, Thailand, for the anti-ζ antibody used in this study. This work was supported by a research and development grant to V.V. by the Faculty of Medicine Siriraj Hospital and MRC Molecular Haematology Unit, Oxford, United Kingdom (to D.R.H.). S.E. and V.V. were supported by the Royal Golden Jubilee PhD program (Grant PHD/0089/2548). S.R. and N.C. received their Masters' thesis funding from tbe Department of Immunology, Siriraj Hospital. V.V. is also supported by a grant from the National Research University, Thailand, through Mahidol University and BIOTEC, Thailand. S.P. received funding from the Landsteiner Foundation for Blood Transfusion Research (LSBR 1040), the Netherlands Scientific Organization (NWO DN 82-301 and ZonMw 912-07-019 and 912-12-128) and the European Union fp7-health-2012 collaborative project THALAMOSS (306201).
Authorship
Contribution: V.V. served as the principle investigator of this study and contributed to case identification, patient care, study design, conducting the study, analysis of the data, and drafting the manuscript; S.E., S.R., N.C., C.F., K.L., H.K., S.B., and J.S. performed laboratory and DNA analysis; M.J., K.S., and V.S.T. took care of patients and collected samples; C.B. and D.S. prepared the manuscript; R.J.G., S.P., and D.R.H. provided support, research direction, and study plan and prepared the manuscript and provided mentorship; and all authors contributed to the data review and provided their comments on the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Vip Viprakasit, Division of Hematology/Oncology, Department of Pediatrics and Thalassemia Center, Faculty of Medicine Siriraj Hospital, Mahidol University, 2 Prannok Road, Bangkoknoi, Bangkok 10700, Thailand; email: vip.vip@mahidol.ac.th.

![Figure 2. Expression profiles of the primary erythroid culture cells from patients with KLF1 mutations. An expression profiling of the α(A) and β(B) globin gene clusters using Agilent tiled oligonucleotide array was performed in P2 erythroid cells compared with normal control. (A) A distinctive higher peak of ζ globin gene expression (dark arrow with gray box highlighted) was observed in the patient (bottom) compared with a normal cDNA from a control sample (top). (B) Two unique upregulated expression peaks of γ-globin (arrow 1) and ε-globin (arrow 2) genes in the patient were shown. Another upregulating expression of noncoding mRNA (arrow 3) was observed at the β-globin locus control region (β-globin locus control region, hypersensitive site 2 [HS-2]), all with dark arrows and gray boxes highlighted. However, the meaning of this observation on the downstream globin gene regulation and hereditary persistence of embryonic globin in the patient remains unclear. There was no significant change on expression of other nearby genes on both clusters. (C) Expression profile of erythroid specific genes from primary erythroid cell samples from 5 patients (P) with KLF1 mutations and normal controls (n = 6), using qPCR and Taqman probe hybridization, showing a marked increase of fetal and embryonic globin mRNA expression (HBZ, ζ-globin gene; HBE, ε-globin gene; HBG, γ-globin gene) in all patients with relatively normal expression of KLF1. This suggests that although most patients carry a single nonsense mutation, which could result in haplo-insufficiency, a missense mutation on the other allele might produce stable transcripts that could be detected at similar levels to normal. Alternatively, our mRNA analysis used might not be sensitive enough to demonstrate a minor perturbation in the level of KLF1 expression. With the exception of BCL11A (B-cell CLL/lymphoma 11A [zinc finger protein] from 1 patient [P2]), there was no change in expression compared with normal controls of other erythroid-specific genes, including CD71, SOX6, ERAF, GYPA, glycophorin A, and EPOR (erythropoietin receptor; supplementary Figure 5).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/123/10/10.1182_blood-2013-09-526087/4/m_1586f2.jpeg?Expires=1769095116&Signature=qL3bLfexB3G2ySLaOtwvK7w5fVEdBGrHNDqew5qewUbs9khm5QyOGnpipnMAa1m2xzGG5ArthYtpAYzO~qT~XxGYLFFkhyugKANiJ0MpWcPZwG1IHvwfecs-Pb2dMOyOrZwhji2c0IkZB0R3AtNwDuATlduH1b5B6yqZAz2B3s~32O~-9rNV5xUROleWkuhB-6QHsDmgLBOvlgn4OKvwmSlCSyopDzhi0islrUak59CzvSqrCB3CQe2j5YWbJG9QFk0fWO7Be2L395bjvqPblgDRQjN5T2TpzXArWuZOptzbdIVhsK07zYU348VEHG1q-vTHF08Y7Xn-yk2Z4A1vEw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal