Key Points
Novel small molecules have been identified that specifically target FVIII.
These small molecules are able to reduce in vitro thrombin formation in full blood.
Abstract
The C domains of coagulation factors V (FV) and VIII (FVIII) are structurally conserved domains and share a common and essential function in membrane binding. In vivo regulation of thrombin formation strongly depends on the expression and regulation of the cofactor activities of FVIII and FV. With this study, we explored the possibility of inhibition of thrombin formation in full blood with small druglike molecules. Such compounds may serve as lead molecules for the development of a new type of orally available coagulation inhibitors that act by blocking the interaction between the C domains of FVIII and the membrane surface. We identified 9 novel molecules that are able to inhibit binding of the FVIII C2 domain to a model membrane by application of a combined ligand-based and target structure–based virtual screening approach that took into account the knowledge of a set of previously identified low-molecular-weight FVIII binders that were, however, not active in full blood. The half-maximal inhibitory concentration values of our newly identified compounds varied from 2.1 to 19.9 µM, of which 7 of 9 molecules did not appreciably inhibit FV membrane binding and were thus specific for FVIII. The most active bioactive compound showed activity in both plasma and in full blood.
Introduction
Despite the almost 60 years of experience with therapeutic anticoagulation, a need still exists for novel types of anticoagulants.1 Well-known adverse effects of anticoagulant therapy include bleeding, unpredictable pharmacokinetics, or immune responses, and several anticoagulants require intravenous administration and monitoring. As such, many initiatives have been undertaken to design still safer anticoagulant drugs with improved risk profiles and patient-friendly routes of administration. Latest developments in the field include the use of orally available small-molecule catalytic site inhibitors of thrombin and factor Xa (FXa).2 The latter new class of molecules is very promising, but the medical community has already noted several well-characterized situations that may restrict their use and thus call on the development of new molecules3,4 such as after a stroke, cerebral bleeding, or in chronic kidney disease.
In our present study, we intended to investigate other molecular mechanisms that could be modulated by small druglike molecules (that could be administered by the oral route after optimization). Also, we decided to focus on anticoagulant molecule candidates that act on the coagulation cofactors factor VIII (FVIII) and/or factor V (FV). The rationale for choosing FVIII and FV as targets is multifaceted and includes the evident analogy with the natural protein C anticoagulant system, the main proteolytic anticoagulant pathway that downregulates thrombin formation though proteolysis of FVIII and FV. Indeed, acting directly on FVIII and/or FV proteins seems pertinent because this would prevent the formation of the procoagulant thrombin and FXa molecules.
The FVIII and FV cofactor proteins are large, multidomain macromolecules that share an A1-A2-B-A3-C1-C2 mosaic protein-domain structure that have been largely underexploited thus far for anticoagulant therapy. Both proteins lack proteolytic activity but enhance the activity of the proteases FXa and factor IXa (FIXa), respectively, by up to 100 000-fold.5,6 Regulation of these cofactor activities is essential for maintenance of hemostatic balance.7-9 To express their procoagulant activities, the activated cofactors must bind to a net negatively charged membrane surface, such as the one provided by the activated thrombocyte in an interaction that requires the presence of the C-terminal C2 domain. Failure to bind to a membrane surface will result in reduced or loss of cofactor activity, which in the case of FVIII and FV is exemplified by both, naturally occurring mutations and/or alloantibodies directed against the C2 domain, which are often associated with a bleeding tendency.10-13
In fact, it is possible to rationally design compounds that act specifically on the C2 domain of FVIII or FV because their 3D structures are known and binding pockets are present that are large enough to bind low-molecular-weight compounds.14-19 Also, it might be easier to obtain specificity on such binding pockets compared with the inhibition of serine protease catalytic sites because these enzymes are present in basically all tissues of the human body.
To our knowledge, only 2 concept studies have been reported so far using this strategy. An experimental high-throughput screening project was performed on FVIII by Spiegel et al,18 and these authors identified ∼10 compounds interacting specifically with the C2 domain of FVIII that impeded membrane binding. Our groups have successfully carried out structure-based virtual screening computations of the C2 domain of FV and found 7 molecules able to interfere with the membrane association of FV. Among them, 4 cross-reacted with FVIII.17 One issue with our previously identified compounds was that they were inactive in human plasma or full blood, in part because of plasma protein binding.
Our present investigation is thus aimed at finding new molecules (optimized compounds and/or novel chemotypes) that could interfere with FVIII membrane binding in blood. We found new molecules using a combination of in silico and in vitro screening experiments, with the best compounds expressing half-maximal inhibitory concentration (IC50) values for the inhibition of FVIII membrane binding of 2.1 to 19.9 µM. The identified compounds may serve as starting points for the development of an entirely new class of anticoagulant molecules, which we name C-domain inhibitors.
Materials and methods
Proteins and reagents
All reagents were of the highest possible grade; details on these are given in the supplemental Methods (available on the Blood Web site).
Druggable pocket definition and virtual screening
The structure of FVIII (1d7p) and FV (1czt) C2 domains were downloaded from the RCSB Protein Data Bank14,15,20 and were used for this study. Details on the structural bioinformatics procedures involved in the definition and preparation of the target, as well as on the virtual screening protocol, are given in supplemental Methods.
SPR-based compound screening
Inhibition of membrane binding was measured via a surface plasmon resonance (SPR)-based assay, performed similar to the SPR-based method that we have described previously.17 We also used SPR to assess the influence of compound C11-10 on the binding between FVIII and von Willebrand factor. Details on the assays are given in the supplemental Methods.
IC50 determination in intrinsic tenase complex
To study the potential of hit molecules to inhibit the activity of the intrinsic tenase complex, we tested their ability to inhibit the formation of FXa by the tenase complex in a system of purified reaction components, essentially as described previously.21 FVIII activity is determined from the rate of FIXa-catalyzed FX activation and FVIII is first preactivated (final concentration 1.5 nM of FVIII) with 2 nM of thrombin for 2 minutes at 37°C followed by the addition of 20 nM of hirudin in buffer I (25 mM of N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.5, 150 mM of NaCl, 0.2 mg/mL of ovalbumin, and 5 mM of CaCl2). Next, the activation mixture was diluted 25-fold into a reaction mixture containing 40 μM of phospholipid vesicles (10:90, DOPS:DOPC, mol:mol) in buffer I. FVIIIa activity reached a stable value after ∼10 minutes of incubation at 37°C, and FX activation was started after 15 minutes of incubation by the addition of FIXa (1 nM) and FX (400 nM). At varying time points, aliquots were taken from the reaction mixture, and the concentration of FXa was determined from the rate of Z-D-Arg-Gly-Arg-p-nitroanilide (S-2765) hydrolysis. By inclusion of varying concentrations of compound molecules in dimethylsulfoxide (DMSO; at 1% final concentration), the inhibition of the tenase complex by test compounds was quantitated by calculation of FXa activities in the presence relative to the FXa activity in the absence of added compounds. We verified that 1% DMSO did not influence the assay outcome.
APTT analysis
To assess the effect of selected compounds on clot formation in plasma, we performed activated partial thromboplastin time (APTT) analysis. To 45 µL of normal pooled platelet-poor plasma (PPP), 5 µL of compound in DMSO was added. Next, 50 µL of Actin FS (Dade Behring, Germany) was added; after another 3-minute incubation, we used 50 µL of CaCl2 (at a final 8.33 mM of concentration) to start coagulation. All incubations were performed for 1 minute at 37°C. Clotting times were recorded in a KC4a micro Coagulometer (Amelung, Germany). We verified that the assay was dependent on the presence of FVIII because addition of a saturating amount of FVIII antibodies prolonged the APTT until >160 seconds.
Activity of hit molecules in human plasma: CAT
Calibrated automated thrombography (CAT) was performed in both PPP and platelet-rich plasma (PRP) as described previously22 while using 1 nM of FIXa, unless otherwise indicated, to trigger coagulation and varying concentrations of compound C11-10. In case of PPP, 4 µM of phospholipid vesicles (20% DOPS, 60% DOPC, 20% DOPE, mol/mol/mol) were added to support coagulation, whereas in PRP 1.50 × 108 platelets/mL were included in the assay (an equivalent of ∼60 µM of lipid vesicles23 ). To prevent contact activation, corn trypsin inhibitor (CTI, 30 μg/mL) was included in all reactions. This CTI concentration completely abolished thrombin generation in plasma triggered with phospholipids and CaCl2 only (no FIXa). Thrombin activity in plasma was monitored continuously with the fluorogenic substrate Z-Gly-Gly-Arg-AMC (BACHEM, Bubendorf, Switzerland). Fluorescence was read in a Fluoroskan Ascent reader (Thermo Labsystems, Helsinki, Finland), and thrombin generation curves were calculated using Thrombinoscope software (Thrombinoscope, Maastricht, The Netherlands).
Effect on clot formation as measured by ROTEM
To study the effect of compound in full blood, we performed rotational elastometry (ROTEM) as a model test that reflects in vivo hemostasis by providing global information on the dynamics of clot development and its stability in a full blood environment. Hemostasis was initiated via the FVIII- and FV-dependent intrinsic route, and total elastograms were recorded. From these, the clotting time, clot formation time, maximal clot firmness, and amplitude at 10 minutes were calculated. We used 1 mg/mL of kaolin and variable amounts of compound, at a 5% percentage of DMSO.
Results
Computational approaches
An overview of the process is presented in supplemental Figure 1. The ChemBridge compound collection was prefiltered (the druglike filter at: http://bioserv.rpbs.univ-paris-diderot.fr/FAF-Drugs/filters.html was used24 ) such as to start with molecules that are predicted orally available and that have the potential to undergo chemical optimization. The resulting collection contained ∼400 000 small molecules.
The first in silico screening approach that we used (ligand-based screening) was carried out over our filtered compound collection. This similarity search strategy aimed at finding close analogs to the previously identified bioactive molecules. We selected 6 query input molecules known to bind to FVIII and/or FV (that can be grouped into 4 structural families). From this in silico screening search, we collected 400 new molecules that could be clustered in 21 different structural families for experimental assays.
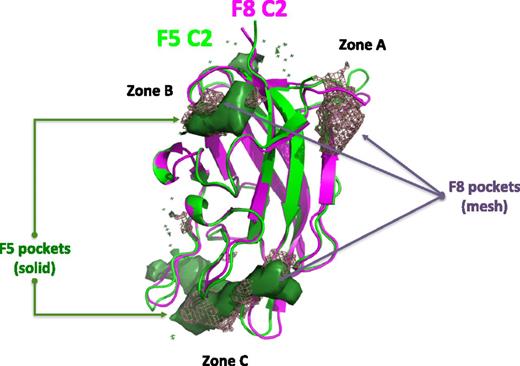
The second in silico strategy applied combines a structure-based screening protocol (ie, docking and scoring) and pharmacophore modeling (ie, a ligand-based approach). The process involved building 4 pharmacophore hypotheses based on known bioactive molecules that were used to screen the “FAF-Drugs filtered” compound collection mentioned above. The top 30 000 molecules were then selected from this first set of computations, resulting in compounds closely related to the queries and in compounds of significant diversity. To reduce the size of the hit list, we decided to use additional constraints coming from the 3D structure of the receptor. Having already explored FV,17 we docked these 30 000 molecules on the C2 domain of FVIII. We first carried out several computations to define the most likely binding pocket(s) in the C2 domains of FVIII and FV. Analysis of the 3D surface of the FVIII C2 domain with several methods, including the recently reported DoGSiteScorer, suggested 3 possible pockets to which a small molecule may bind (zones A-C, Figure 1). On FV, a total of 2 “druggable” binding sites were identified (zones B-C). Thus, only FVIII appears to have a binding zone A; indeed, the 3D structure of the 2 domains shows some differences in a loop region in this area (Figure 1). Overall, zones B and C are present on both FV and FVIII. Experimentally, it is known that a compound inhibiting FV membrane interaction also inhibits the binding of a monoclonal antibody that interacts in the overall area of zone C.17 Furthermore, very poor binding scores and binding poses were obtained when attempting to dock our 6 query molecules in zones A and B. This finding suggests that the area of zone C is the most likely binding pocket. Certainly, this region contains several amino acids and loops known to be involved in direct membrane interaction25 and would be consistent with our previous electron microscopy data showing that the tip of the C2 domain (and part of the tip of the C1 domain) inserts into the phospholipids.26
In silico analysis of small molecule binding pockets on the surface of the C2 domain of human FVIII and FV. Ribbon diagram presentation of the C2 domain of FVIII (magenta) and FV (green). The consensus from pocket detection programs is shown here, illustrating the presence of 3 potential locations for pockets that may bind small molecules on the surface of the C2 domains: in zones A, B, and C. Zone A was found only in the FVIII structure. The overall shape of the pockets identified is indicated in a mesh representation for FVIII and a solid surface for FV.
In silico analysis of small molecule binding pockets on the surface of the C2 domain of human FVIII and FV. Ribbon diagram presentation of the C2 domain of FVIII (magenta) and FV (green). The consensus from pocket detection programs is shown here, illustrating the presence of 3 potential locations for pockets that may bind small molecules on the surface of the C2 domains: in zones A, B, and C. Zone A was found only in the FVIII structure. The overall shape of the pockets identified is indicated in a mesh representation for FVIII and a solid surface for FV.
Structure-based virtual screening computations with Surflex27 were thus performed, focusing on the zone C area of the FVIII C2 domain. After docking the 30 000 molecules resulting from our pharmacophore screening, the top 1000 binding energy scores were selected, and visual inspection allowed us to reject 377 molecules because of unlikely poses or very high internal strain energies. A list of 623 molecules was then generated. Among these molecules, we noticed that 23 molecules had already been identified by the similarity search method described above. Thus, finally, 600 molecules were selected. By clustering these molecules using the same parameters as above for the ligand-based screening runs, we obtained a total of 150 clusters, of which some contain only 2 to 3 family members and 27 singletons.
We then compiled a final list of 1000 molecules from our different in silico computations that were purchased. With the clustering approach used (see the “Materials and methods” section), these molecules clustered in 158 chemical families (containing 2-35 members) and 8 singletons.
SPR-based compound screening
A total of 26 mixes of the 250 mixtures of 4 compounds tested were identified as containing hits, and these mixtures were further deconvoluted to individually test the compounds in a similar SPR setup. The analysis was performed at 2.5 µM of concentration of compound to establish which of the 4 compounds could be classified as a hit. This yielded a total of 14 compounds with an inhibitory activity of >20% at 2.5 µM. Of these compounds, 5 were excluded because they appeared to bind directly to the membrane surface.
IC50 determination
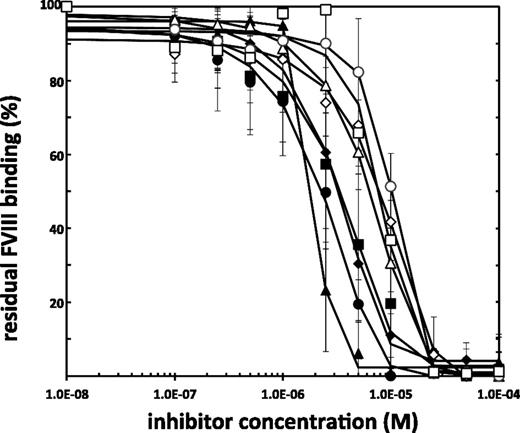
For each of the remaining 9 hit molecules, the IC50 was determined in the SPR-based assay described above. To this end, varying concentrations of compound were included in the assay, and the resulting final levels of FVIII binding were expressed relative to the binding in the absence of compound. Results for these analyses are shown in Figure 2. Each data point represents the average of at least 3 measurements. IC50 values for the hit molecules are given in Table 1. To verify that the compounds were specifically targeting the C2 domain of FVIII, we used the SPR-based method as described for FVIII, but we injected the purified isolated FVIII-C2 domain (400 nM of final concentration) instead, to obtain IC50 values for inhibition of the binding of the FVIII-C2 domain to lipid membranes. By applying the intrinsic tenase assay described above, we further determined IC50 values for inhibition of the tenase complex by the 9 hit molecules (Table 1). We illustrate the dose-dependency of the inhibition of the intrinsic tenase in supplemental Figure 2, whereby the nonabrupt and saturable inhibition is observed for compound C11-10.
IC50 determination of hit molecules as selected by the SPR-based assay. Increasing concentrations of compound are incubated with 4 nM of FVIII, which is next injected in a Biacore T100. The residual binding of FVIII to a 10/90 M/M PS/PC membrane surface in the presence of small-molecule inhibitors (flowcell 2), corrected for binding to a 100% PC (flowcell 1), relative to the FVIII binding in the absence of small-molecule inhibitors is given here. Lines indicate a nonlinear fit of the data points (sigmoidal dose-response fit). Shown are averages ± standard deviation for at least 2 independent experiments per point. ◆, A11-8;▲, D7-5; ●, H5-8; ▪, C11-10; ⋄, C8-4; △, D11-5; ○, C8-5; □, B7-5.
IC50 determination of hit molecules as selected by the SPR-based assay. Increasing concentrations of compound are incubated with 4 nM of FVIII, which is next injected in a Biacore T100. The residual binding of FVIII to a 10/90 M/M PS/PC membrane surface in the presence of small-molecule inhibitors (flowcell 2), corrected for binding to a 100% PC (flowcell 1), relative to the FVIII binding in the absence of small-molecule inhibitors is given here. Lines indicate a nonlinear fit of the data points (sigmoidal dose-response fit). Shown are averages ± standard deviation for at least 2 independent experiments per point. ◆, A11-8;▲, D7-5; ●, H5-8; ▪, C11-10; ⋄, C8-4; △, D11-5; ○, C8-5; □, B7-5.
IC50 values determined by SPR-based and tenase activity assays
| Compound ID . | ChemBridge ID . | IC50 for inhibition of FVIII-membrane binding, µM (mean) . | IC50 for inhibition of tenase activity, µM (mean) . | IC50 for inhibition of C2 domain-membrane binding, µM (mean) . |
|---|---|---|---|---|
| A11-8* | 5992471 | 3.3 | 9.2 | 3.9 |
| D7-5 | 5651006 | 2.1 | 26.9 | 2.4 |
| H5-8 | 6844287 | 2.7 | 3.5 | 47 |
| C11-10 | 7687521 | 3.5 | 4.2 | 4.6 |
| C8-4 | 5898659 | 9.6 | 5.6 | 52.5 |
| D11-5 | 5941257 | 6.7 | 10.1 | 22.9 |
| D6-7 | 6656941 | 19.9 | 59.7 | 33.7 |
| C8-5 | 5740628 | 10.6 | 18.5 | 33.6 |
| B7-5 | 5731791 | 8.1 | 38.6 | 20.2 |
| Compound ID . | ChemBridge ID . | IC50 for inhibition of FVIII-membrane binding, µM (mean) . | IC50 for inhibition of tenase activity, µM (mean) . | IC50 for inhibition of C2 domain-membrane binding, µM (mean) . |
|---|---|---|---|---|
| A11-8* | 5992471 | 3.3 | 9.2 | 3.9 |
| D7-5 | 5651006 | 2.1 | 26.9 | 2.4 |
| H5-8 | 6844287 | 2.7 | 3.5 | 47 |
| C11-10 | 7687521 | 3.5 | 4.2 | 4.6 |
| C8-4 | 5898659 | 9.6 | 5.6 | 52.5 |
| D11-5 | 5941257 | 6.7 | 10.1 | 22.9 |
| D6-7 | 6656941 | 19.9 | 59.7 | 33.7 |
| C8-5 | 5740628 | 10.6 | 18.5 | 33.6 |
| B7-5 | 5731791 | 8.1 | 38.6 | 20.2 |
All values are averages of at least 2 dose-response curves (for SPR-based methods) and of triplicates (tenase activity assay).
Protein specificity determination by SPR
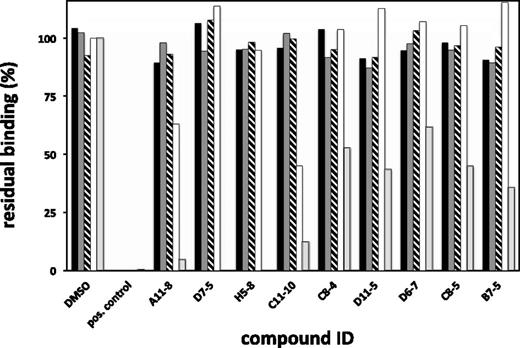
To address the selectivity of our compounds, we investigated the ability of hit molecules to inhibit membrane binding of several selected membrane-binding coagulation proteins, other than FVIII, via an SPR-based assay, as described above and as reported previously by our group.17 We injected FV (40 nM), prothrombin (1 µM), FIXa (1 µM), and FXa (250 nM), at a fixed concentration of compound (10 µM). Figure 3 illustrates the amount of binding, as expressed relative to membrane binding of the respective proteins in the absence of compound. For comparison, the averaged values from the FVIII curves, at 10 µM of compound, have been included in this figure.
Binding specificity of hit compounds as determined by direct-binding analysis using SPR. FIXa (black bars), FXa (dark gray bars), prothrombin (striped), or FV (white) were tested for their ability to bind to a 10/90 PS/PC phospholipid membrane in the presence of fixed concentrations of 10 µM of hit molecules. For comparison also, the binding of FVIII (in light gray) at this concentration was included. Residual binding was calculated by division of the binding signal in the presence of compound over the signal in the absence of compound (DMSO only), while correcting for binding to 100% PC membranes. Positive controls were included in the assay to ascertain the correctness of the assay outcome. As a positive control runs for FIXa, FXa and prothrombin were performed in the presence of 20 mM of EDTA, whereas for FV and FVIII, an inhibitory antibody directed against the C2 domain was used at a concentration 0.25 mg/mL. Data shown are averages of at least 2 individual measurements, with the standard error of the mean being <15% for all values.
Binding specificity of hit compounds as determined by direct-binding analysis using SPR. FIXa (black bars), FXa (dark gray bars), prothrombin (striped), or FV (white) were tested for their ability to bind to a 10/90 PS/PC phospholipid membrane in the presence of fixed concentrations of 10 µM of hit molecules. For comparison also, the binding of FVIII (in light gray) at this concentration was included. Residual binding was calculated by division of the binding signal in the presence of compound over the signal in the absence of compound (DMSO only), while correcting for binding to 100% PC membranes. Positive controls were included in the assay to ascertain the correctness of the assay outcome. As a positive control runs for FIXa, FXa and prothrombin were performed in the presence of 20 mM of EDTA, whereas for FV and FVIII, an inhibitory antibody directed against the C2 domain was used at a concentration 0.25 mg/mL. Data shown are averages of at least 2 individual measurements, with the standard error of the mean being <15% for all values.
Activity of hit molecules in human plasma: APTT
In a previous study, we have observed the binding to albumin of several of our FV C2 domain binders.17 Therefore, to ascertain if our new hit molecules were active in a plasma environment, we performed an APTT screen at a high concentration of 333 µM of compound. A significant prolongation of the APTT was observed only for compound C11-10 and, to a lesser extent, A11-8, whereas the other compounds did not significantly induce a change of APTT time (data not shown). Together with the earlier determined relatively low IC50 values for C11-10, we selected this compound for further analysis in plasma. To verify the specificity of the inhibition, we performed a titration experiment in which we included increasing amounts of C11-10 (0-500 µM of C11-10) at constant DMSO concentration (1.67%) in an APTT test in normal pooled plasma. Figure 4A illustrates the dose-response curve for the prolongation of the APTT by addition of C11-10. At 500 µM of C11-10, the recorded time exceeded the maximal time of the instrument (999 seconds) (time point not shown in Figure 4A). Given that the APTT at high C11-10 concentrations exceeds that of when a saturating amount of FVIII inhibitory was added, we conclude that under these conditions not only FVIII, but also FV inhibition (as was noted from the SPR data presented above in Figure 3), contributes to the prolongation of the clotting time.
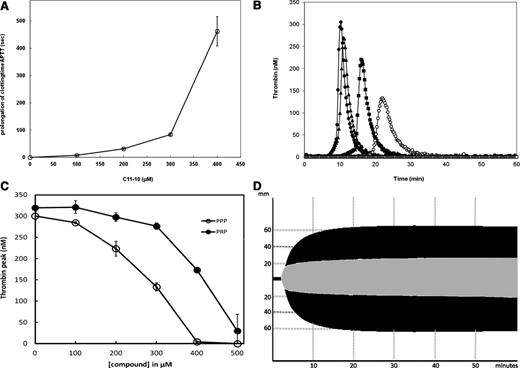
Characterization of anticoagulant properties of compoundC11-10 (7687521) in human plasma and full blood. (A) Prolongation of clotting time in platelet-poor human pooled plasma by C11-10. APTT was performed as described in the “Materials and methods” section, with varying amounts of C11-10 present (from 0-500 µM). A value for the prolongation of the APTT at 500 µM could not be obtained because this time exceeds 999 seconds and is thus not indicated in this graph. Data points are averages ± standard deviation from 3 measurements. (B) Influence of compound C11-10 in thrombin formation in PPP. Thrombin generation was triggered by 1 nM of FIXa in PPP in the presence of 4 µM of phospholipid vesicles (20% DOPS, 60% DOPC, 20% DOPE, mol/mol/mol), 30 µg/mL of CTI, and varying concentrations of compound C11-10 from 0 to 500 µM. Various concentrations of C11-10 are indicated by symbols: ● (0 µM), ▲ (100 µM), ▪ (200 µM), ○ (300 µM), △ (400 µM), and □ (500 µM). At 400 and 500 µM of C11-10 no thrombin was formed during the time of the experiment. (C) Inhibition of thrombin formation in PPP and PRP by compound C11-10. CAT was performed as described in the “Materials and methods” section, using PPP (○) or PRP (●). The thrombin peaks (in nM) are shown as a function of the final concentration of C11-10 present. Averages ± standard deviation are indicated, n = 3. (D) Influence on hemostasis in full blood by C11-10 as determined by ROTEM. ROTEM analysis was performed in full blood of a normal healthy individual in the absence (black graph) and presence (gray graph) of 500 µM of C11-10 after intrinsic trigger (1 mg/mL of kaolin) and a final concentration of 11.8 mM of CaCl2 added.
Characterization of anticoagulant properties of compoundC11-10 (7687521) in human plasma and full blood. (A) Prolongation of clotting time in platelet-poor human pooled plasma by C11-10. APTT was performed as described in the “Materials and methods” section, with varying amounts of C11-10 present (from 0-500 µM). A value for the prolongation of the APTT at 500 µM could not be obtained because this time exceeds 999 seconds and is thus not indicated in this graph. Data points are averages ± standard deviation from 3 measurements. (B) Influence of compound C11-10 in thrombin formation in PPP. Thrombin generation was triggered by 1 nM of FIXa in PPP in the presence of 4 µM of phospholipid vesicles (20% DOPS, 60% DOPC, 20% DOPE, mol/mol/mol), 30 µg/mL of CTI, and varying concentrations of compound C11-10 from 0 to 500 µM. Various concentrations of C11-10 are indicated by symbols: ● (0 µM), ▲ (100 µM), ▪ (200 µM), ○ (300 µM), △ (400 µM), and □ (500 µM). At 400 and 500 µM of C11-10 no thrombin was formed during the time of the experiment. (C) Inhibition of thrombin formation in PPP and PRP by compound C11-10. CAT was performed as described in the “Materials and methods” section, using PPP (○) or PRP (●). The thrombin peaks (in nM) are shown as a function of the final concentration of C11-10 present. Averages ± standard deviation are indicated, n = 3. (D) Influence on hemostasis in full blood by C11-10 as determined by ROTEM. ROTEM analysis was performed in full blood of a normal healthy individual in the absence (black graph) and presence (gray graph) of 500 µM of C11-10 after intrinsic trigger (1 mg/mL of kaolin) and a final concentration of 11.8 mM of CaCl2 added.
Activity of hit molecules in human plasma: CAT
Figure 4B illustrates the result from CAT performed in PPP, at varying C11-10 concentrations (0-500 µM). At 400 and 500 µM of C11-10, thrombin formation was completely blocked and no residual signal could be observed. Under these conditions, thrombin formation depends on FVIII because in a control experiment performed with FVIII deficient plasma, we did not measure any appreciable thrombin formation (see also supplemental Figure 3, compare to TF-triggered CAT). To assess the possible influence of platelets in plasma to influence the inhibition by C11-10, we repeated the CAT with PRP, the results of which have been summarized in Figure 4C. For comparison, the observed peak heights in PPP, as calculated from the graphs in Figure 4B, are shown as well.
Effect on clot formation in full blood
ROTEM analysis was performed to study the effect of C11-10 in full blood. Figure 4D illustrates a typical curve, which exemplifies the attenuation of hemostasis in full blood by C11-10. Further illustration of this effect can be seen in supplemental Table 1, which summarizes the results from the ROTEM analysis. In all cases, increasing amounts of C11-10 results in a slight shortening of the clotting time (time from start to when the waveform reaches 2 mm above baseline), whereas the clot formation time (time from 2 mm above baseline to 20 mm above baseline) is clearly prolonged, and the maximal clot firmness and amplitude at 10 minutes are significantly reduced.
In silico analysis of the 9 hits identified in our present study
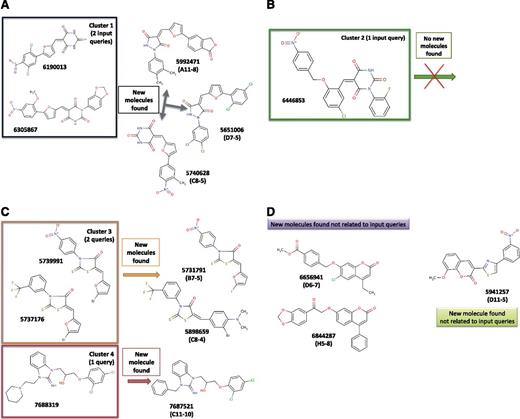
It appears that the molecules identified in the present study are, in general, structurally closely related to our query compounds; this observation is, to some extent, as expected with the strategy used (Figure 5A-D). The best compound for its action in full blood is the molecule named here C11-10, which results from a simple similarity search protocol. Compound C11-10 is structurally very similar to, yet different from, compound 7688319 (Figure 5C) (see below).
Query molecules and hit compounds grouped in structural families. The 6 starting molecules known to interact with FV and/or FVIII (the queries) from previous studies can be grouped into 4 families (named here as clusters 1, 2, 3, and 4). We used these starting structures to search new molecules that could be structurally similar to the queries or relatively different (ie, both types of molecules are relevant). If we group the 9 active molecules found in our present study, we notice that 3 new compounds are related to cluster 1 (this one contains 2 members) (A), 2 new molecules are close to 2 query molecules grouped here under the name of cluster 3, whereas 1 new molecule is similar to 1 query compound (named here as cluster 4) (C). We found 3 other new molecules that are structurally unrelated to our starting queries. These 3 compounds can be grouped structurally into 2 families, one containing 2 members (6656941 and 6844287) and the other, only 1 member (5941257) (D). Cluster 2 did not result in the identification of new molecules (B). The reasoning behind such organization of the molecular structures is that it helps to rationalize the activity and is important for future optimization.
Query molecules and hit compounds grouped in structural families. The 6 starting molecules known to interact with FV and/or FVIII (the queries) from previous studies can be grouped into 4 families (named here as clusters 1, 2, 3, and 4). We used these starting structures to search new molecules that could be structurally similar to the queries or relatively different (ie, both types of molecules are relevant). If we group the 9 active molecules found in our present study, we notice that 3 new compounds are related to cluster 1 (this one contains 2 members) (A), 2 new molecules are close to 2 query molecules grouped here under the name of cluster 3, whereas 1 new molecule is similar to 1 query compound (named here as cluster 4) (C). We found 3 other new molecules that are structurally unrelated to our starting queries. These 3 compounds can be grouped structurally into 2 families, one containing 2 members (6656941 and 6844287) and the other, only 1 member (5941257) (D). Cluster 2 did not result in the identification of new molecules (B). The reasoning behind such organization of the molecular structures is that it helps to rationalize the activity and is important for future optimization.
Compound C11-10 displays physicochemical properties that are very acceptable at this stage of the drug discovery process (supplemental Figure 4) and could be used as starting point for further chemical optimization.
Redocking the best compound into the C2 domain
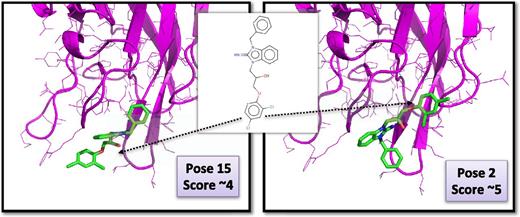
To investigate further the most likely position of compound C11-10 at the surface of the C2 domain of FVIII, we redocked this molecule in the area of zone C using advanced parameters not compatible with fast virtual screening while allowing exploration of up to ∼9 Å around this predicted binding pocket.
The 2 most likely poses are presented in Figure 6 (from the 40 poses generated). With these orientations, we note in each case hydrophobic and aromatic contacts and 3H bonds between the molecule and the C2 domain. Redocking of C11-10 was also performed on the FV C2 domain, and several poses could be considered as equivalent to the ones obtained for FVIII, potentially explaining the observed cross-reactivity with FV that we noted for this molecule (data not shown).
Redocking of compoundC11-10 in the FVIII C2 domain. Shown are the ribbon diagrams and the side chains of the C2 domain of human FVIII (in magenta) in the area of the binding pocket. Compound C11-10 is in a stick representation and the 2 most likely positions as obtained after extensive docking and scoring are displayed. Other poses were obtained (>40) but were rejected when they could not make favorable hydrogen bonds and/or hydrophobic/aromatic contacts with the proteins or when the ligand had very high internal strain energy.
Redocking of compoundC11-10 in the FVIII C2 domain. Shown are the ribbon diagrams and the side chains of the C2 domain of human FVIII (in magenta) in the area of the binding pocket. Compound C11-10 is in a stick representation and the 2 most likely positions as obtained after extensive docking and scoring are displayed. Other poses were obtained (>40) but were rejected when they could not make favorable hydrogen bonds and/or hydrophobic/aromatic contacts with the proteins or when the ligand had very high internal strain energy.
Discussion
Applying the approach described here that combined ligand-based and structure-based virtual screening computations, we were able to retrieve 9 active molecules that inhibit the binding of FVIII to a membrane surface with an IC50 between 2.1 and 19.9 µM, and with 4 compounds having an IC50 of <3.5 µM (Figure 2; Table 1).
Of the 9 molecules identified, 7 were specific for FVIII, whereas 2 molecules (A11-8 and C11-10) also inhibited the binding of FV to a membrane layer. These 9 molecules can be grouped into 5 families, and they are linked to our input queries, although 3 molecules (grouped into 2 clusters) can be considered as new chemicals (Figure 5). The hit rate found in our study (0.9%; ie, 9 actives while testing 1000 molecules) is largely underestimated, and it should be noted that a large number of molecules, which can equally be described as “hit,” with intermediate activities were among the selections made via the virtual methods. This is evident when analyzing the results of the initial screening of compound mixes because after a first screen at 62.5 µM (of each compound in the mixture of 4), we found 79% of the mixes to present an inhibitory activity of >70%. Because we were interested in finding relatively high-affinity molecules, we decided to perform the screening at 2.5 µM (for each compound in the mixture), which then reduced the percentage of actives to 10.4%.
Importantly, the molecule referred here as C11-10 proved to possess appreciable anticoagulant activity when tested in plasma or in full blood. To the best of our knowledge, this is the first time that a small molecule acting by blocking FVIII/FV is able to inhibit blood coagulation and hemostasis in plasma containing a normal amount of 1.50 × 108 platelets/mL and in whole blood. Although C11-10 had IC50 values for inhibition of FVIII, the FVIII C2 domain and the FXase complex that were in the low micromolar region (Table 1), larger concentrations were required to affect coagulation in plasma-containing reaction systems. Control experiments showed that binding of C11-10 to albumin is a likely contributor to this difference (supplemental Figure 5). Interestingly, C11-10 did not prohibit the binding of FVIII to von Willebrand factor, suggesting that this compound will not affect the circulatory half-life of FVIII (supplemental Figure 6).
It seems that part of the efficacy of C11-10 could be due to the fact that it acts on both FVIII and FV. Indeed, given the overall 40% sequence identity between the C2 domains of FVIII and FV, we attribute this cross-reactivity to the structural similarity of the target structures. To analyze further this hypothesis, we investigated potential “druggable” pockets at the surface of the C2 domain of both cofactors using several algorithms (supplemental Methods), and only 2 similar binding regions (zones B and C) could be identified. Among them, the relatively conserved zone C (on FVIII and FV) appears to be the only one allowing favorable interactions with our bioactive compounds (Figures 1 and 6) and could explain the dual inhibition of a molecule such as C11-10. On the other hand, this compound appears specific for FVIII and FV and does not act on FIXa, FXa, or prothrombin.
C11-10 is structurally similar to a molecule identified in a previous study that was cocrystallized with the C2 domain of FVIII (PDB file 3HNB28 ) in a position that is different from the most likely position determined by our computations. In the crystal structure the ligand is in a region where no binding sites were predicted (supplemental Figure 7) and where the compound appears to somewhat lie on a very flat surface. Some aromatic and hydrogen bond interactions are noticed in the experimental structure between the ligand and the protein but could not be reproduced by 3 different docking methods (Surflex, SwissDock, Molegro Virtual docker). Additional investigations are therefore needed because at ∼90% of the time, a small molecule binds to the top 3 to 5 in silico–predicted binding regions.29
Most low-molecular-weight compounds bind to ∼4 to 9 targets in average30 (in general, not with the same affinity), and we also noticed that our best molecule could eventually bind to some “off-target” using prediction servers such as SEA31 and ChemProt.32 Interestingly, the same compound has been tested on 8 different targets/phenotypes not involved in coagulation, as seen by searching the chEMBL database,33 but the affinities were relatively low (∼50 µM), or the molecule was only found active without affinity measurements. This finding suggests that after optimization, C11-10 could clearly become an interesting lead molecule.
With our current work, we propose that efficient in vivo anticoagulation can be achieved with C-domain inhibitors, a potential novel class of small anticoagulant molecules that inhibit the membrane binding of FV and FVIII.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We greatly acknowledge Dr J.W.N. Heemskerk for his assistance in the thromboelastometry experiments.
This work was supported by grants from the Netherlands Organisation for Scientific Research (91107006) (G.A.F.N.), the Transnational University Limburg (G.A.F.N), the Cardiovascular Research Institute Maastricht, and the Institut National de la Santé et de la Recherche Médicale (INSERM) Institute.
Authorship
Contribution: G.A.F.N., M.K., P.H.K., A.W., S.W., R.S., and O.S. performed experiments; G.A.F.N., J.V., and B.O.V. analyzed results and created the figures; and G.A.F.N. and B.O.V. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gerry Nicolaes, Department of Biochemistry, Cardiovascular Research Institute Maastricht, Maastricht University, PO Box 616, 6200 MD Maastricht, The Netherlands; e-mail: g.nicolaes@maastrichtuniversity.nl.