Key Points
Mer tyrosine kinase is aberrantly expressed in ∼30% of pediatric pre–B-ALL patients, including most patients with an E2A-PBX1 translocation.
Mer inhibition decreased B-ALL cell survival signal transduction, caused chemosensitization, and prolonged survival in a xenograft model.
Abstract
Acute lymphoblastic leukemia (ALL) is currently treated with an intense regimen of chemotherapy yielding cure rates near 85%. However, alterations to treatment strategies using available drugs are unlikely to provide significant improvement in survival or decrease therapy-associated toxicities. Here, we report ectopic expression of the Mer receptor tyrosine kinase in pre–B-cell ALL (B-ALL) cell lines and pediatric patient samples. Inhibition of Mer in B-ALL cell lines decreased activation of AKT and MAPKs and led to transcriptional changes, including decreased expression of antiapoptotic PRKCB gene and increase in proapoptotic BAX and BBC3 genes. Further, Mer inhibition promoted chemosensitization, decreased colony-forming potential in clonogenic assays, and delayed disease onset in a mouse xenograft model of leukemia. Our results identify Mer as a potential therapeutic target in B-ALL and suggest that inhibitors of Mer may potentiate lymphoblast killing when used in combination with chemotherapy. This strategy could reduce minimal residual disease and/or allow for chemotherapy dose reduction, thereby leading to improved event-free survival and reduced therapy-associated toxicity for patients with B-ALL. Additionally, Mer is aberrantly expressed in numerous other malignancies suggesting that this approach may have broad applications.
Introduction
Cancer is the leading cause of disease-related death among children, and acute lymphoblastic leukemia (ALL) is the most common pediatric malignancy. B-precursor ALL (B-ALL), the most common pediatric ALL subtype, can be further classified by chromosomal translocation.1 One common B-ALL chromosomal rearrangement is t(1;19),2 a fusion of the E2A and PBX1 transcription factors,3 which promotes oncogenesis through altered regulation of gene expression.
While chemotherapy has dramatically increased cure rates,4 significant risk of short- and long-term toxicities (neurocognitive sequelae, cardiovascular dysfunction, secondary malignancies, infertility) persist. The incidence of severe late effects is ∼25%.5,6 Furthermore, survival rates for children with relapsed ALL remain poor.7 Novel approaches are needed to increase efficacy and/or reduce toxicity.
Molecularly targeted agents have advanced the treatment of certain pediatric ALLs. Use of BCR-ABL tyrosine kinase inhibitors in Philadelphia chromosome-positive ALL dramatically increased event-free survival from 35% to 80%.8 FLT-3 tyrosine kinase inhibitors are undergoing trials in pediatric MLL-rearranged ALL.9,10 The early success of targeting proteins expressed by cancer cells or aberrant intracellular signaling pathways supports the development of additional targeted agents in the treatment of pediatric leukemia to further increase therapeutic efficacy.
Within the hematopoietic lineages, Mer receptor tyrosine kinase is expressed on dendritic cells, macrophages, natural killer cells, and megakaryocytes/platelets.11 Mer is not expressed in normal lymphocytes12-14 but is a highly upregulated gene in E2A-PBX1+ pre–B-ALL.1 Ectopic expression is seen in pediatric T-cell ALL.13 Transgenic Mer expression in mice results in the development of T-cell and biphenotypic lymphoblastic leukemia and lymphoma.14 Mer activates antiapoptotic signaling proteins, including AKT and ERK1/2,15-17 and promotes tumorigenic phenotypes in several other tumor types, including non–small-cell lung cancer,18 melanoma,19 and acute myeloid leukemia.20
Here, we present data demonstrating increased Mer protein in E2A-PBX1+ and other cytogenetic subgroups of B-ALL. By using both in vitro and in vivo models, we provide compelling evidence that Mer inhibition attenuates prosurvival and proliferation signaling, enhances apoptosis in response to chemotherapy, and significantly delays the onset of leukemia in a xenograft mouse model. These data validate Mer as a novel B-ALL therapeutic target, suggesting that therapies targeting Mer may enhance chemotherapeutic response and thereby allow dose reduction and decreased toxicity.
Materials and methods
Patient samples and cell culture
Deidentified diagnostic bone marrow or peripheral blood samples were obtained from the Children’s Oncology Group or Children’s Hospital Colorado Tissue Bank after written informed consent in accordance with the Declaration of Helsinki. Experiments conformed to regulatory standards as approved by the Colorado Multiple Institutional Review Board. REH, RCH-ACV, UOCB1, Hal01, Nalm6, and RS411 cell lines were generously provided by Lia Gore, MD (University of Colorado Denver). The 697 cell line was from the German Collection of Microorganisms and Cell Cultures. Cell lines were maintained in RPMI medium (Sigma-Aldrich) supplemented with 10% fetal bovine serum and penicillin/streptomycin (cRPMI). Parental and knockdown ALL cell line identities were confirmed by using short tandem repeat microsatellite loci analysis.
Flow cytometric analysis of cell surface proteins
Archival patient blood or bone marrow samples were evaluated for Mer surface expression using a phycoerythrin (PE)-conjugated mouse-anti-human Mer antibody. Prospective patient samples were analyzed by flow cytometry within 24 hours of sample collection. Mouse immunoglobulin G1 isotype control and PE-labeled secondary antibodies defined background staining such that ≤2% of lymphoblast populations (CD19+/CD45+) were identified as expressing Mer. This gate was then applied to a sample from the same patient stained with Mer primary and PE-labeled secondary antibodies to determine the percentage of lymphoblasts expressing Mer. Cell lines were similarly analyzed. Hematopoietic progenitor populations from C57Bl/6 Mer+/+, Mer−/−, and Mer transgenic (MerTg) murine bone marrow were analyzed by flow cytometry as previously described.21 See supplemental Methods for additional details (available on the Blood Web site).
Immunoblot analysis
Cells were cultured in serum-free medium (Gas6 treated) or cRPMI (chemotherapy treated) for 3 to 4 hours and then treated with 200 nM recombinant human Gas6 (R&D Systems) or chemotherapeutics (Sigma-Aldrich) for the indicated times and concentrations. Whole-cell lysates were prepared, and proteins were resolved on tris(hydroxymethyl)aminomethane (Tris)-glycine sodium dodecyl sulfate polyacrylamide gel electrophoresis gels (Invitrogen) and transferred onto polyvinylidine difluoride membranes. Membranes were blocked in Tris-buffered saline with 0.1% Tween-20 containing 5% milk (see supplemental Methods for additional details).
Real-time quantitative RT-PCR
Total RNA was isolated from patient samples using a spin column method (RNeasy Plus Mini Kit; Qiagen). Real-time reverse-transcription polymerase chain reaction (RT-PCR) analysis was performed by using TaqMan Universal PCR Master Mix with No AmpErase UNG (Applied Biosystems) (see supplemental Methods for details). Threshold cycle values were normalized to the 18S ribosomal RNA internal control, and analysis was performed as previously described.22 A twofold difference in RNA concentration per cycle was assumed for calculation of fold-change values.
RNA-seq and data analysis
After treatment with Gas6 or methotrexate, RNA was extracted from the 697 cell line as above. A complementary DNA library was constructed and sequenced on a HiSeq-2000 (Illumina) at the University of Colorado Anschutz Medical Campus Genomics and Microarray Core. On average, 50 million single-end 100-bp sequencing reads per sample were obtained. RNA-seq analysis was performed as previously described23,24 by using Tophat/Cufflinks workflow.25 To determine the differentially expressed genes, cuffdiff (with false-discovery rate <0.001, fold-change >2, and both FPKM [fragments per kb of transcript per million fragments mapped] values >1) was used. Differentially expressed genes were analyzed in the National Institutes of Health Database for Annotation, Visualization and Integrated Discovery (NIH DAVID)26 for functional and pathway enrichment.
Knockdown of Mer via RNA interference
Lentiviral vectors (pLKO.1) containing short hairpin RNA (shRNA) sequences targeting Mer (shMer1, Oligo ID: TRCN0000000862 and shMer4, Oligo ID: TRCN0000000865) or green fluorescent protein (GFP) (shControl, Catalog No. RHS4459) were obtained from Open Biosystems. The GFP vector expresses an shRNA targeting the GFP protein, which is not expressed by human cells and thus serves as a nonsilencing control. Replication-incompetent virus particles were generated and B-ALL cells were transduced as previously described.18 Clonal populations of REH and 697 shMer cells were isolated by fluorescence-assisted single-cell sorting.
Chemosensitivity assays
To assess apoptosis, cells were cultured in the presence or absence of chemotherapies for 48 hours. Harvested cells were washed, resuspended in phosphate-buffered saline containing 1 µM YO-PRO-1 iodide (Invitrogen) and 1.5 µM propidium iodide (PI) (Invitrogen), and incubated on ice for 15 to 20 minutes. Fluorescence was detected and analyzed by using an FC 500 flow cytometer with CXP data analysis software (Beckman Coulter). For determination of recovery after chemotherapy, cells were cultured as above for 48 hours. Harvested cells were washed twice, and 1.5 × 105 viable cells per mL were cultured in cRPMI for 6 days. Viable lymphoblast count was determined by trypan blue exclusion by using a Cedex XS Analyzer (Innovatis).
Clonogenic assays
Cells were plated in triplicate at a density of 500 cells per mL in ColonyGel Human Base Medium methylcellulose (Reach Bio) supplemented with cRPMI per manufacturer’s instructions. Plates were incubated for 10 days at 37°C with 5% CO2, and colonies were counted by using a GelCount automated colony counter (Oxford Optronix) after staining with 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT; Sigma-Aldrich) reagent.
Xenograft mouse model of leukemia
Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice were purchased from Jackson Laboratories. Experiments conformed to the relevant regulatory standards as approved by the University of Colorado Institutional Animal Care and Use Committee. Wild-type or shRNA-containing cells were injected via tail vein into NSG mice. Animals were monitored daily and euthanized upon development of physical signs of significant leukemia burden (>5% weight loss, abnormal posture, decreased activity, labored breathing, and/or hind limb paralysis).
Statistical analysis
Statistical analyses were performed by using GraphPad Prism software v5.01 (GraphPad Software). Flow cytometric apoptosis assays were analyzed by using a paired Student t test. All remaining results were analyzed by using an unpaired Student t test to compare shMer knockdowns to shControls. Kaplan-Meier survival curves were analyzed by using a Mantel-Cox test. Results were considered significant at P < .05.
Results
Mer tyrosine kinase is ectopically expressed in pediatric B-ALL
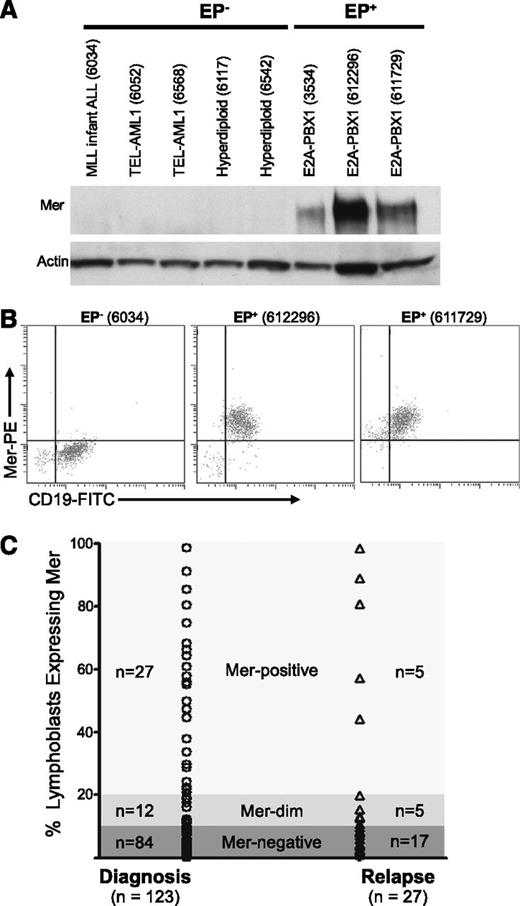
Archived pediatric B-ALL samples from newly diagnosed patients were evaluated for Mer messenger RNA (mRNA) (via RT-PCR) and protein expression (via immunoblot and flow cytometric analyses). All E2A-PBX1+ samples (n = 16) expressed Mer protein (Figure 1A-B; supplemental Table 1; data not shown). In contrast, 11 of 12 E2A-PBX1− samples did not express Mer RNA transcript and/or protein. Four E2A-PBX1+ and 6 E2A-PBX1− samples were analyzed by quantitative RT-PCR (qRT-PCR). Expression of Mer mRNA and protein correlated in all cases (supplemental Table 1). The E2A-PBX1+ samples exhibited a 10- to 205-fold increase in Mer mRNA transcript relative to 5 E2A-PBX1− samples, which expressed negligible levels of Mer mRNA (data not shown). Although 11 of 12 samples from newly diagnosed E2A-PBX1− patients did not express Mer, Mer was detected in 7 of 7 human B-ALL cell lines tested, including all 5 E2A-PBX1− cell lines; 3 of these 5 cell lines expressed Mer at levels comparable to E2A-PBX1+ cell lines (supplemental Figure 1).
Mer is expressed in diagnostic samples derived from patients with B-ALL. Twenty-eight diagnostic bone marrow samples from patients with B-cell ALL were analyzed by (A) immunoblot and/or (B) flow cytometry for the expression of Mer protein. (A) Immunoblot analysis of Mer protein (∼180 kDa) in representative samples of non–E2A-PBX1 (EP−) B-ALL (lanes 1-5) and E2A-PBX1+ (EP+) B-ALL (lanes 6-8). Patient sample ID is shown in parentheses for comparison; the complete data set is listed in supplemental Table 1. The membrane was stripped and reprobed with anti-actin (∼43 kDa) antibody to confirm similar loading of total protein. (B) Representative flow cytometry profiles for one EP− B-ALL sample (6034) and two EP+ B-ALL samples (612296 and 611729) are shown. All three samples expressed the CD19 B-lineage marker whereas only the EP+ samples exhibited Mer staining. (C) Mer expression was determined by flow cytometry in a prospective analysis of samples collected at diagnosis or relapse from patients with B-ALL. Each symbol represents data collected from a single sample. Approximately 32% of patients at diagnosis and 37% of patients at relapse express Mer on leukemic blasts (Mer positive + Mer dim).
Mer is expressed in diagnostic samples derived from patients with B-ALL. Twenty-eight diagnostic bone marrow samples from patients with B-cell ALL were analyzed by (A) immunoblot and/or (B) flow cytometry for the expression of Mer protein. (A) Immunoblot analysis of Mer protein (∼180 kDa) in representative samples of non–E2A-PBX1 (EP−) B-ALL (lanes 1-5) and E2A-PBX1+ (EP+) B-ALL (lanes 6-8). Patient sample ID is shown in parentheses for comparison; the complete data set is listed in supplemental Table 1. The membrane was stripped and reprobed with anti-actin (∼43 kDa) antibody to confirm similar loading of total protein. (B) Representative flow cytometry profiles for one EP− B-ALL sample (6034) and two EP+ B-ALL samples (612296 and 611729) are shown. All three samples expressed the CD19 B-lineage marker whereas only the EP+ samples exhibited Mer staining. (C) Mer expression was determined by flow cytometry in a prospective analysis of samples collected at diagnosis or relapse from patients with B-ALL. Each symbol represents data collected from a single sample. Approximately 32% of patients at diagnosis and 37% of patients at relapse express Mer on leukemic blasts (Mer positive + Mer dim).
A prospective flow cytometric analysis of bone marrow or blood samples from pediatric B-ALL patients was performed at diagnosis or relapse to determine cell surface expression of Mer. Samples that had >20% cells with fluorescence intensity above background were defined as “Mer-positive,” and samples with 10% to 20% cells above background were defined as “Mer-dim.” Samples with <10% above background were termed “Mer-negative,” although these samples may still have low-level Mer expression. Of 123 samples from newly diagnosed patients, 32% (39 of 123) expressed Mer on leukemic blasts (Figure 1C). Mer expression in B-ALL patient samples at relapse was similar; 37% (10 of 27) expressed Mer. Of the E2A-PBX1+ samples in this prospective analysis, the majority (78%; 7 of 9) expressed Mer protein (supplemental Table 2). No correlation between Mer expression and patient age, gender, or additional specific molecular rearrangement was observed.
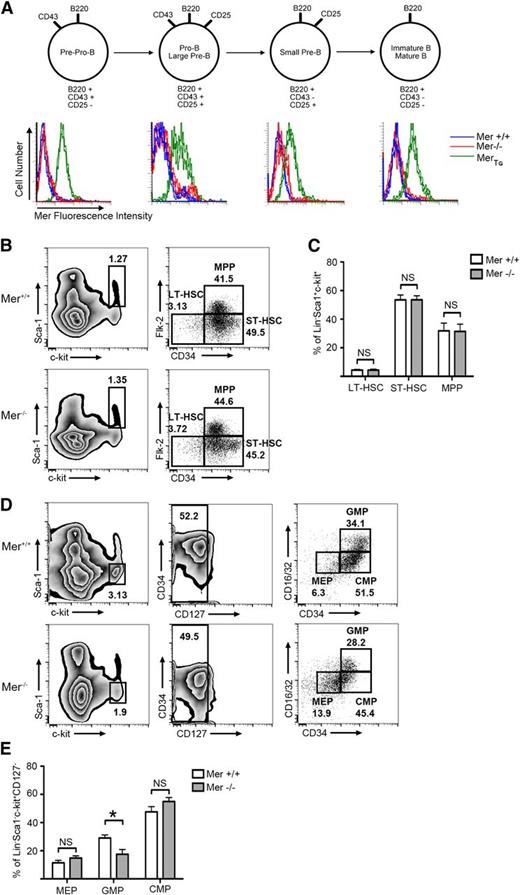
We have previously reported that Mer is not expressed in normal mature T and B lymphocytes, or at any stage of T-cell development.12-14 Similarly, flow cytometric analysis demonstrated that Mer was not detected on the surface of B cells at any stage of development (Figure 2A). To understand the impact that therapeutic Mer inhibition may have on bone marrow progenitors, we analyzed the effects of Mer absence on progenitor populations. Analysis by flow cytometry of marrow derived from Mer−/− and Mer+/+ mice revealed no difference in the percentage of hematopoietic stem cell or multipotent progenitor populations (Figure 2B-C). A decrease in the percentage of granulocyte-macrophage progenitors in Mer knockout mice was observed (18% ± 3% in Mer−/−; 29% ± 2% in Mer+/+), although no difference in other myeloid progenitors was noted (Figure 2D-E).
Mer is not expressed on the surface of murine B-lineage cells. Bone marrow was harvested from wild-type (Mer+/+), Mer knockout (Mer−/−), or MerTg C57Bl/6 mice. (A) The remaining lymphocytes were stained with antibodies that bind to the CD45R (B220), CD43, and CD25 surface receptors to classify B cells into various stages of maturation, as depicted in the diagram. Cells were also stained with anti-mouse Mer antibody to detect surface expression of Mer. The histograms demonstrate that Mer is not expressed on the surface of B cells from wild-type mice at any stage of maturation. B cells from Mer−/− or MerTg mice were used as negative and positive controls, respectively. Representative data from 4 independent experiments are shown. (B) Representative flow cytometry plots from Mer+/+ and Mer−/− bone marrow samples identifying the long-term hematopoietic stem cell (LT-HSC; Lin−Sca1+ckit+CD34−Flk2−), short-term hematopoietic stem cell (ST-HSC; Lin−Sca1+ckit+CD34+Flk2−), and multipotent progenitor (MPP; Lin−Sca1+ckit+CD34+Flk2+) populations. (C) Quantification of the LT-HSC, ST-HSC, and MPP populations from Mer+/+ and Mer−/− bone marrow (n = 4). (D) Representative flow cytometry plots identifying the megakaryocyte-erythroid progenitors (MEPs; Lin−Sca1+ckit+CD127−CD34−CD16/32−), granulocyte-macrophage progenitors (GMPs; Lin−Sca1+ckit+CD127−CD34+CD16/32+), and common myeloid progenitors (CMPs; Lin−Sca1+ckit+CD127−CD34+CD16/32−) from Mer+/+ and Mer−/− bone marrow. (E) Quantification of the MEP, GMP, and CMP populations from Mer+/+ and Mer−/− bone marrow (n = 5). All graphs show the mean ± standard error of the mean (SEM) from at least two independent experiments. *P < .05. NS, not significant.
Mer is not expressed on the surface of murine B-lineage cells. Bone marrow was harvested from wild-type (Mer+/+), Mer knockout (Mer−/−), or MerTg C57Bl/6 mice. (A) The remaining lymphocytes were stained with antibodies that bind to the CD45R (B220), CD43, and CD25 surface receptors to classify B cells into various stages of maturation, as depicted in the diagram. Cells were also stained with anti-mouse Mer antibody to detect surface expression of Mer. The histograms demonstrate that Mer is not expressed on the surface of B cells from wild-type mice at any stage of maturation. B cells from Mer−/− or MerTg mice were used as negative and positive controls, respectively. Representative data from 4 independent experiments are shown. (B) Representative flow cytometry plots from Mer+/+ and Mer−/− bone marrow samples identifying the long-term hematopoietic stem cell (LT-HSC; Lin−Sca1+ckit+CD34−Flk2−), short-term hematopoietic stem cell (ST-HSC; Lin−Sca1+ckit+CD34+Flk2−), and multipotent progenitor (MPP; Lin−Sca1+ckit+CD34+Flk2+) populations. (C) Quantification of the LT-HSC, ST-HSC, and MPP populations from Mer+/+ and Mer−/− bone marrow (n = 4). (D) Representative flow cytometry plots identifying the megakaryocyte-erythroid progenitors (MEPs; Lin−Sca1+ckit+CD127−CD34−CD16/32−), granulocyte-macrophage progenitors (GMPs; Lin−Sca1+ckit+CD127−CD34+CD16/32+), and common myeloid progenitors (CMPs; Lin−Sca1+ckit+CD127−CD34+CD16/32−) from Mer+/+ and Mer−/− bone marrow. (E) Quantification of the MEP, GMP, and CMP populations from Mer+/+ and Mer−/− bone marrow (n = 5). All graphs show the mean ± standard error of the mean (SEM) from at least two independent experiments. *P < .05. NS, not significant.
shRNA knockdown of Mer inhibits ligand-stimulated and chemotherapy-induced prosurvival signaling in B-ALL
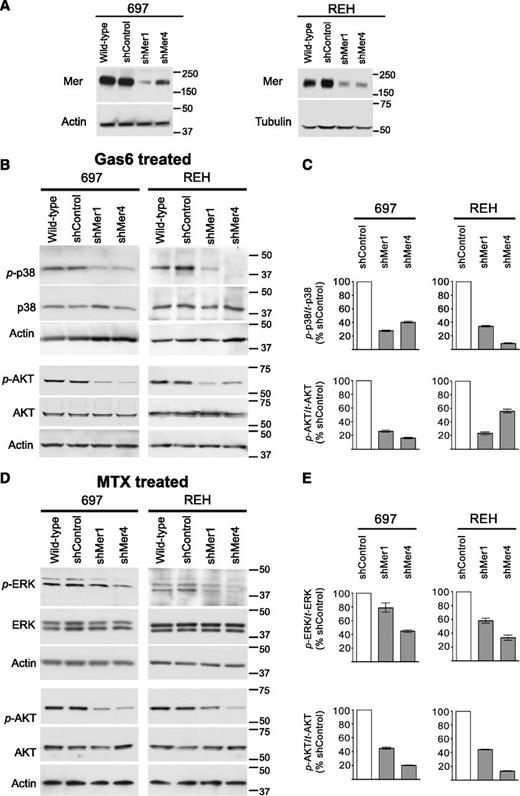
Two Mer-expressing B-ALL cells lines, 697 (E2A-PBX1+) and REH (E2A-PBX1−), were selected for further investigation. Two independent shRNA constructs (shMer1, shMer4) were used to inhibit Mer expression in these cell lines, and stable clonal populations were derived. Immunoblot analysis indicated robust knockdown of Mer protein expression in 697 and REH shMer1 and shMer4 clones relative to wild-type cells or cells transduced with a nonsilencing control shRNA (shControl) (Figure 3A). Flow cytometric analysis confirmed decreased Mer cell surface expression relative to shControl cells in both 697 (77% reduction for shMer1; 64% reduction for shMer4) and REH (58% reduction for shMer1; 57% reduction for shMer4) knockdown clones (supplemental Figure 2).
Mer knockdown decreases intracellular survival and proliferation signaling. Wild-type cells were infected with lentiviral particles containing 1 of 2 independent shRNA constructs targeting Mer (shMer1, shMer4) or GFP (shControl) as a nonsilencing control. (A) Mer knockdown was confirmed by immunoblot analysis of whole-cell lysate. Equal amounts of total protein were analyzed to qualitatively demonstrate reduced expression of Mer (∼180 kDa). Blots were stripped and reprobed with anti-actin antibody (∼43 kDa) or anti-tubulin antibody (∼55 kDa) to confirm similar protein loading of 697 and REH lysates, respectively. (B) Wild-type, shControl, and Mer knockdown (shMer1, shMer4) 697 and REH cells were incubated in serum-free medium for 3 hours prior to stimulation with 200 nM Gas6 for 10 minutes or (D) cultured in cRPMI and treated with 30 nM methotrexate (MTX) for 1 hour. Whole-cell lysates were prepared and phosphorylated (denoted by p-) and total proteins were detected. Immunoblots representative of 3 independent experiments are shown. Blots were stripped and reprobed with anti-actin antibody (∼43 kDa) to confirm similar protein loading. (C) Ratio of phosphorylated p38 MAPK and AKT to total protein level was decreased in shMer1 and shMer4 cells relative to shControl cells following stimulation with Gas6. (E) Ratio of phosphorylated ERK1/2 and AKT was decreased in shMer1 and shMer4 cells relative to shControl cells following treatment with MTX.
Mer knockdown decreases intracellular survival and proliferation signaling. Wild-type cells were infected with lentiviral particles containing 1 of 2 independent shRNA constructs targeting Mer (shMer1, shMer4) or GFP (shControl) as a nonsilencing control. (A) Mer knockdown was confirmed by immunoblot analysis of whole-cell lysate. Equal amounts of total protein were analyzed to qualitatively demonstrate reduced expression of Mer (∼180 kDa). Blots were stripped and reprobed with anti-actin antibody (∼43 kDa) or anti-tubulin antibody (∼55 kDa) to confirm similar protein loading of 697 and REH lysates, respectively. (B) Wild-type, shControl, and Mer knockdown (shMer1, shMer4) 697 and REH cells were incubated in serum-free medium for 3 hours prior to stimulation with 200 nM Gas6 for 10 minutes or (D) cultured in cRPMI and treated with 30 nM methotrexate (MTX) for 1 hour. Whole-cell lysates were prepared and phosphorylated (denoted by p-) and total proteins were detected. Immunoblots representative of 3 independent experiments are shown. Blots were stripped and reprobed with anti-actin antibody (∼43 kDa) to confirm similar protein loading. (C) Ratio of phosphorylated p38 MAPK and AKT to total protein level was decreased in shMer1 and shMer4 cells relative to shControl cells following stimulation with Gas6. (E) Ratio of phosphorylated ERK1/2 and AKT was decreased in shMer1 and shMer4 cells relative to shControl cells following treatment with MTX.
Previous reports demonstrate that Mer stimulation in various cell types leads to phosphorylation of AKT, mTOR, ERK1/2, and p38 MAPK, resulting in reduced apoptosis.15,17,18 Treatment with Gas6, a known Mer ligand,18,27 resulted in phosphorylation of AKT (S473) and p38 (Thr180/Tyr182) in wild-type and shControl cells. In contrast, ligand-dependent phosphorylation of AKT and p38 were diminished in shMer1 and shMer4 cells (Figure 3B-C). Phosphorylation of ERK1/2 (T202/Y204) and mTOR (S2448) did not increase in response to Gas6 (data not shown).
Additionally, we treated REH and 697 cells with methotrexate, a chemotherapeutic agent commonly used in the treatment of ALL. Compared with shControl cells, Mer knockdown cells demonstrated decreased phosphorylation of AKT and ERK1/2 (Figure 3D-E), although p38 and mTOR phosphorylation were unchanged (data not shown).
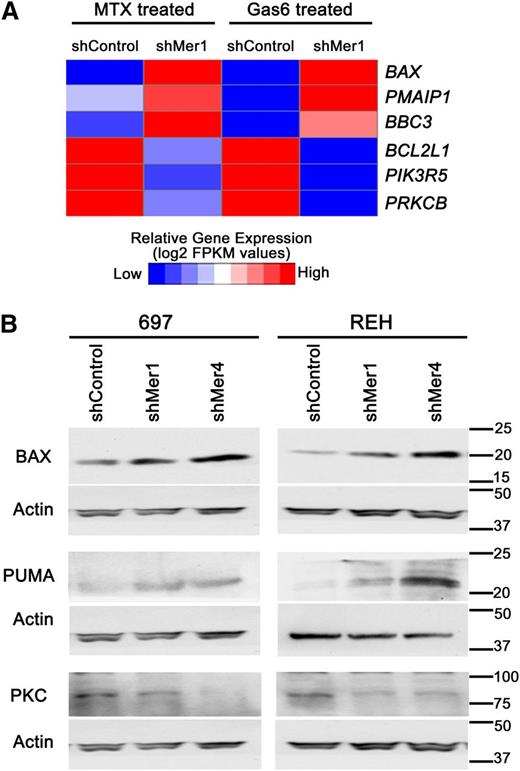
To identify novel targets downstream of Mer, alterations in gene expression in Mer knockdown B-ALL cells relative to control cells after treatment with Gas6 or methotrexate were determined by RNA sequencing. This analysis revealed decreased expression of prosurvival genes and increased expression of proapoptotic genes in Mer knockdown cells. Specifically, the prosurvival BCL2L1 (BCL-XL), PIK3R5 (phosphatidyl inositol 3 kinase [PI3K]), and PRKCB (protein kinase C, PKC) were all found to be downregulated when Mer was inhibited (Figure 4A). Furthermore, the proapoptotic BAX, PMAIP1 (NOXA), and BBC3 (PUMA) were all found to be upregulated when Mer was inhibited. We validated the upregulation of BAX and PUMA, and downregulation of PKC protein levels (Figure 4B) in Mer knockdown cells relative to control cells.
Mer knockdown attenuates pro-survival gene regulation and increases pro-apoptotic gene expression. (A) After treatment with 200 nM Gas6 for 10 minutes or 30 nM methotrexate for 1 hour, shMer1 knockdown and shControl 697 cells were harvested and RNA was extracted. RNA sequencing was performed revealing increased mRNA expression of proapoptotic genes BAX (BAX), PMAIP1 (NOXA), and BBC3 (PUMA) and decreased expression of prosurvival genes BCL2L1 (BCL-XL), PIK3R5 (PI3K), and PRKCB (PKC) in shMer knockdown cells relative to control cells after treatment with Gas6 or methotrexate. (B) Immunoblot analysis of BAX, PUMA, and PKC protein levels in shMer knockdown and shControl 697 cells treated with Gas6 or methotrexate demonstrating protein levels concordant with the altered gene expression profiles observed in panel A.
Mer knockdown attenuates pro-survival gene regulation and increases pro-apoptotic gene expression. (A) After treatment with 200 nM Gas6 for 10 minutes or 30 nM methotrexate for 1 hour, shMer1 knockdown and shControl 697 cells were harvested and RNA was extracted. RNA sequencing was performed revealing increased mRNA expression of proapoptotic genes BAX (BAX), PMAIP1 (NOXA), and BBC3 (PUMA) and decreased expression of prosurvival genes BCL2L1 (BCL-XL), PIK3R5 (PI3K), and PRKCB (PKC) in shMer knockdown cells relative to control cells after treatment with Gas6 or methotrexate. (B) Immunoblot analysis of BAX, PUMA, and PKC protein levels in shMer knockdown and shControl 697 cells treated with Gas6 or methotrexate demonstrating protein levels concordant with the altered gene expression profiles observed in panel A.
Inhibition of Mer in combination with chemotherapy promotes apoptosis
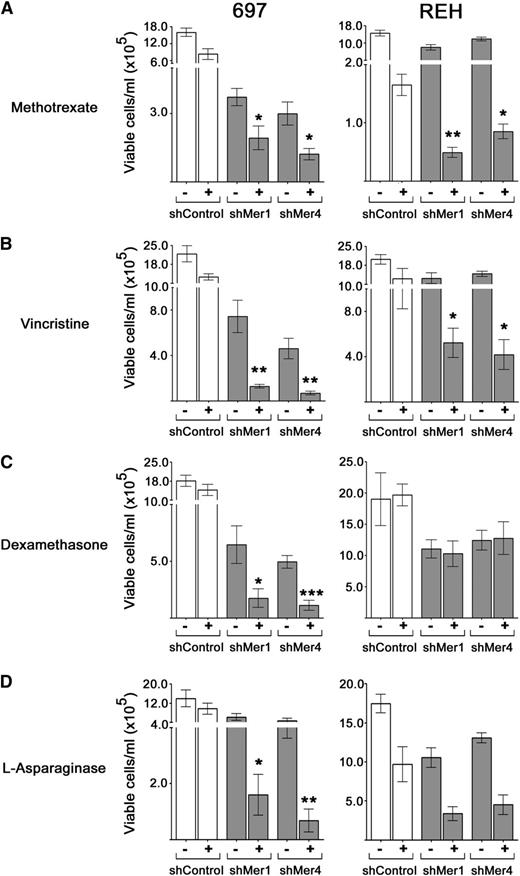
Given the observed modulation of survival and proliferation signaling in 697 and REH Mer knockdown cell lines, we investigated whether Mer inhibition enhanced chemosensitivity. After 48-hour treatment with various chemotherapeutics, B-ALL cells were stained with YO-PRO and PI and analyzed by flow cytometry. Cells undergoing early apoptosis take up YO-PRO, and cells undergoing later apoptosis or necrosis take up both YO-PRO and PI (Figure 5A). In both 697 and REH cells, Mer knockdown increased apoptosis in response to treatment with methotrexate or vincristine (Figure 5A-C). Moreover, Mer inhibition interacted synergistically with both drugs to mediate leukemia cell killing (supplemental Table 3). Mer knockdown also sensitized 697 cells to treatment with dexamethasone and l-asparaginase mediated by a synergistic induction of cell death. A similar trend was observed in REH cells treated with l-asparaginase. In contrast, Mer inhibition had no effect on sensitivity to dexamethasone in REH cells (Figure 5D-E). Mer inhibition did not affect cell death in response to treatment with doxorubicin, etoposide, and 6-mercaptopurine (data not shown).
Inhibition of Mer expression increases cell death and promotes apoptotic signaling pathway activation in response to chemotherapy. Apoptotic and dead cells were identified by flow cytometric analysis of cells stained with YO-PRO-1 iodide and PI. (A) Representative 697 flow cytometry profiles after treatment with methotrexate (MTX) are shown. The percentages of cells in early apoptosis (YO-PRO-1 positive, right lower quadrant), cells in later stages of apoptosis and necrosis (PI positive, upper quadrants), and live cells (left lower quadrant) are shown. Cultures of 697 and REH shControl, shMer1, and shMer4 cells were exposed to (B) 30 nM methotrexate, (C) 0.2 nM vincristine (VINC), (D) 50 nM (697) or 10 µm (REH) dexamethasone (DEX), or (E) 3 IU/mL l-asparaginase (L-ASP) or equivalent vehicle (phosphate-buffered saline [PBS] for methotrexate and vincristine; ethanol for dexamethasone and l-asparaginase) for 48 hours. Drug treated denoted as “+”, and vehicle treated denoted as “−”. Mean values and SEs derived from at least 3 independent experiments are shown. In response to methotrexate and vincristine, both REH and 697 Mer knockdown cells (shMer1 and shMer4) exhibited significant synergistic increases in the total percentage of apoptotic and dead cells compared to drug-treated shControl cells. In response to dexamethasone and l-asparaginase treatment, 697 cells exhibit significant induction of apoptosis. REH shMer cells exhibited a trend toward increased sensitivity in response to l-asparaginase, although there was no change in response to dexamethasone. *P < .05; **P < .01. (F) Whole-cell lysates were prepared from 697 and REH wild-type, shControl, shMer1, and shMer4 cells treated with methotrexate for 48 hours, and PARP, caspase 3, and caspase 8 were detected by immunoblot analysis. Representative 697 immunoblots are shown.
Inhibition of Mer expression increases cell death and promotes apoptotic signaling pathway activation in response to chemotherapy. Apoptotic and dead cells were identified by flow cytometric analysis of cells stained with YO-PRO-1 iodide and PI. (A) Representative 697 flow cytometry profiles after treatment with methotrexate (MTX) are shown. The percentages of cells in early apoptosis (YO-PRO-1 positive, right lower quadrant), cells in later stages of apoptosis and necrosis (PI positive, upper quadrants), and live cells (left lower quadrant) are shown. Cultures of 697 and REH shControl, shMer1, and shMer4 cells were exposed to (B) 30 nM methotrexate, (C) 0.2 nM vincristine (VINC), (D) 50 nM (697) or 10 µm (REH) dexamethasone (DEX), or (E) 3 IU/mL l-asparaginase (L-ASP) or equivalent vehicle (phosphate-buffered saline [PBS] for methotrexate and vincristine; ethanol for dexamethasone and l-asparaginase) for 48 hours. Drug treated denoted as “+”, and vehicle treated denoted as “−”. Mean values and SEs derived from at least 3 independent experiments are shown. In response to methotrexate and vincristine, both REH and 697 Mer knockdown cells (shMer1 and shMer4) exhibited significant synergistic increases in the total percentage of apoptotic and dead cells compared to drug-treated shControl cells. In response to dexamethasone and l-asparaginase treatment, 697 cells exhibit significant induction of apoptosis. REH shMer cells exhibited a trend toward increased sensitivity in response to l-asparaginase, although there was no change in response to dexamethasone. *P < .05; **P < .01. (F) Whole-cell lysates were prepared from 697 and REH wild-type, shControl, shMer1, and shMer4 cells treated with methotrexate for 48 hours, and PARP, caspase 3, and caspase 8 were detected by immunoblot analysis. Representative 697 immunoblots are shown.
Consistent with the results of the flow apoptosis assays, shMer1 and shMer4 knockdown cell lines demonstrated increased poly adenosine diphosphate ribose polymerase (PARP), caspase 3, and caspase 8 cleavage in response to treatment with methotrexate relative to shControl cells treated with methotrexate (Figure 5F).
To further assess the combination of Mer inhibition and chemotherapy, cells were treated with chemotherapeutics for 48 hours and then allowed to recover in normal culture conditions without chemotherapy. Consistent with the apoptosis assays, both 697 and REH Mer knockdown cells exhibited greater sensitivity to methotrexate and vincristine, with significantly fewer proliferating cells posttreatment relative to shControl cells (Figure 6A-B). Similarly, 697 knockdown cells were more sensitive to dexamethasone and l-asparaginase, whereas REH knockdown cells demonstrated a trend toward increased sensitivity in response to treatment with l-asparaginase but not dexamethasone (Figure 6C-D). Of note, in the setting of addition of chemotherapy vehicle, Mer inhibition alone often diminished the repopulating capabilities of these B-ALL cells.
Inhibition of Mer expression results in increased chemosensitivity in B-ALL cells. (A) Mer knockdown (shMer1, shMer4) and shControl 697 and REH cells were treated with 30 nM methotrexate (+) or equivalent concentration of PBS (−) (vehicle control) for 48 hours, then washed, resuspended in cRPMI medium in equal numbers, and cultured for 6 days. Similarly, cells were treated with (B) 0.2 nM vincristine or PBS, (C) 50 nM dexamethasone for 697 and 10 µm dexamethasone for REH or ethanol, and (D) 3 IU/mL l-asparaginase or ethanol. At the end of 6 days, the concentration of viable cells was determined. Mean values and SEs derived from at least 3 independent experiments are shown. (A-B) Rebound growth of 697 and REH cells after methotrexate or vincristine treatment was abrogated by Mer inhibition compared with shControl cells. (C) Rebound growth of 697 cells after treatment with dexamethasone was abrogated by Mer inhibition although Mer inhibition did not affect steroid resistance in REH cells. (D) Rebound growth of 697 cells after treatment with l-asparaginase was abrogated by Mer inhibition. A similar trend was observed for REH cells, although both knockdowns were not statistically significant. *P < .05; **P < .01; ***P < .001.
Inhibition of Mer expression results in increased chemosensitivity in B-ALL cells. (A) Mer knockdown (shMer1, shMer4) and shControl 697 and REH cells were treated with 30 nM methotrexate (+) or equivalent concentration of PBS (−) (vehicle control) for 48 hours, then washed, resuspended in cRPMI medium in equal numbers, and cultured for 6 days. Similarly, cells were treated with (B) 0.2 nM vincristine or PBS, (C) 50 nM dexamethasone for 697 and 10 µm dexamethasone for REH or ethanol, and (D) 3 IU/mL l-asparaginase or ethanol. At the end of 6 days, the concentration of viable cells was determined. Mean values and SEs derived from at least 3 independent experiments are shown. (A-B) Rebound growth of 697 and REH cells after methotrexate or vincristine treatment was abrogated by Mer inhibition compared with shControl cells. (C) Rebound growth of 697 cells after treatment with dexamethasone was abrogated by Mer inhibition although Mer inhibition did not affect steroid resistance in REH cells. (D) Rebound growth of 697 cells after treatment with l-asparaginase was abrogated by Mer inhibition. A similar trend was observed for REH cells, although both knockdowns were not statistically significant. *P < .05; **P < .01; ***P < .001.
Mer expression enhances colony-forming potential in B-ALL
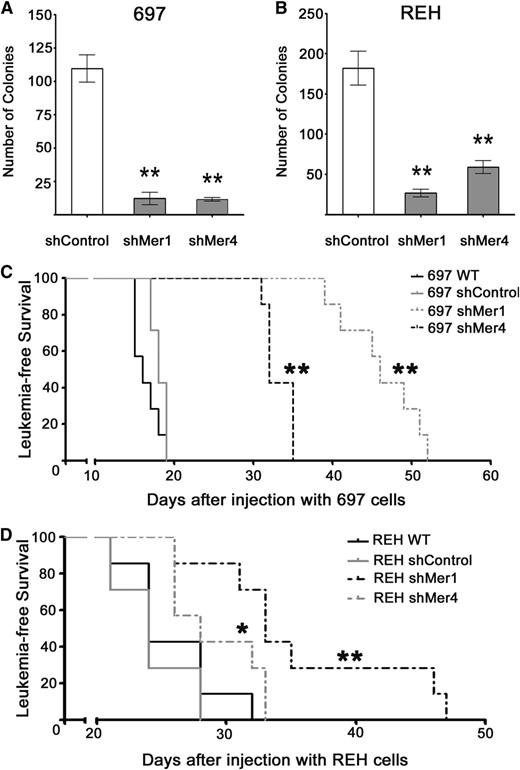
We determined the effects of Mer inhibition on growth in methylcellulose, which may more closely mimic the conditions encountered by leukemia cells in vivo. Control and Mer knockdown B-ALL cells were plated in equal number in methylcellulose, and colony numbers were determined after 10 days. In both 697 and REH cultures, Mer inhibition significantly impaired colony formation ability (Figure 7A-B). Mer knockdown 697 cells demonstrated a 90% decrease in colony-forming units compared with shControl (12 ± 4.6 for shMer1, 11.4 ± 1.4 for shMer4, 110 ± 10.2 for shControl; Figure 7A). Mer knockdown REH cells demonstrated a 67% to 85% decrease in colony-forming units compared with shControl (27 ± 4.7 for shMer1, 59 ± 8.0 for shMer4, 182 ± 21.0 for shControl; Figure 7B). These data suggest that Mer may mediate a leukemogenic mechanism in addition to cell survival and antiapoptosis. Note that both 697 and REH cells were more sensitive to Mer inhibition in the colony-forming assay than they were in liquid cultures (Figures 5 and 6), suggesting that Mer may mediate functions important for tumor-promoting interactions within the microenviroment.
Mer inhibition is sufficient to significantly decrease colony formation and inhibit leukemogenesis in a mouse xenograft model of pre–B-ALL. Equal numbers of (A) 697, (B) REH shControl, and Mer knockdown (shMer1, shMer4) cells were cultured in methylcellulose for 10 days. Mer knockdown cells formed significantly fewer colonies than shControl cells. Mean values and SEs derived from at least 3 independent experiments are shown. (C-D) NSG mice (n = 7 per group) were injected intravenously with 2 × 106 (697) or 3 × 106 (REH) wild-type, shControl, shMer1, or shMer4 cells. (C) Comparison of survival curves for 697 xenografts revealed a significant increase in leukemia-free survival with Mer inhibition (median survival: 46 days for shMer1, 32 days for shMer4) compared with shControl xenografts (median survival: 18 days). No significant difference was observed between 697 xenografts injected with shControl and wild-type (median survival: 16 days) cells. (D) Comparison of survival curves for REH xenografts revealed a significant increase in leukemia-free survival with Mer inhibition (median survival: 33 days for shMer1, 28 days for shMer4) compared with shControl xenografts (median survival: 24 days). No significant difference was observed between REH xenografts injected with shControl and wild-type (median survival 24 days) cells. *P < .05; **P < .01.
Mer inhibition is sufficient to significantly decrease colony formation and inhibit leukemogenesis in a mouse xenograft model of pre–B-ALL. Equal numbers of (A) 697, (B) REH shControl, and Mer knockdown (shMer1, shMer4) cells were cultured in methylcellulose for 10 days. Mer knockdown cells formed significantly fewer colonies than shControl cells. Mean values and SEs derived from at least 3 independent experiments are shown. (C-D) NSG mice (n = 7 per group) were injected intravenously with 2 × 106 (697) or 3 × 106 (REH) wild-type, shControl, shMer1, or shMer4 cells. (C) Comparison of survival curves for 697 xenografts revealed a significant increase in leukemia-free survival with Mer inhibition (median survival: 46 days for shMer1, 32 days for shMer4) compared with shControl xenografts (median survival: 18 days). No significant difference was observed between 697 xenografts injected with shControl and wild-type (median survival: 16 days) cells. (D) Comparison of survival curves for REH xenografts revealed a significant increase in leukemia-free survival with Mer inhibition (median survival: 33 days for shMer1, 28 days for shMer4) compared with shControl xenografts (median survival: 24 days). No significant difference was observed between REH xenografts injected with shControl and wild-type (median survival 24 days) cells. *P < .05; **P < .01.
Inhibition of Mer expression increases survival in a mouse xenograft B-ALL model
To validate Mer as a novel therapeutic target in vivo, we evaluated the oncogenicity of control and Mer knockdown 697 and REH cells in a mouse xenograft model. Wild-type, shControl and shMer cells were injected intravenously into NSG mice and survival was monitored. Median survival for animals injected with wild-type or shControl 697 cells was 16 and 18 days, respectively (Figure 7C). In contrast, median survival was significantly greater for animals transplanted with shMer1 (46 days; P < .01) or shMer4 (32 days; P < .01) knockdown cells. Median survival of mice injected with wild-type or shControl REH cells was 24 days, whereas median survival for mice transplanted with shMer1 or shMer4 cells was 33 days (P < .01) and 28 days (P < .05), respectively (Figure 7D). Thus, Mer inhibition significantly delays the development and/or progression of leukemia in vivo.
Discussion
In these studies, we demonstrated that Mer is ectopically expressed in B-ALL. In cell lines and newly diagnosed patients with B-ALL, Mer mRNA1 and protein expression is associated with presence of the E2A-PBX1 fusion protein. However, Mer protein was also detected in E2A-PBX1− B-ALL cell lines and pediatric patient samples. Of the five E2A-PBX1− cell lines that expressed Mer, four were established from patients at relapse and one from a patient for whom induction therapy failed,28-30 suggesting that Mer expression may be more prevalent in chemotherapy resistant and/or recurrent B-ALL. Our current prospective analysis of Mer expression in lymphoblasts isolated from pediatric patients indicates a similar frequency of Mer expression at diagnosis and relapse, although the sample size is insufficient to fully analyze the relation between Mer expression and prognosis; these studies are ongoing. This is, however, the first study in which Mer expression was analyzed prospectively in a large number of patients with B-ALL, and it demonstrates feasibility of flow cytometric detection of Mer as a strategy to identify patients who might benefit from treatment with a Mer inhibitor.
The functional significance of this study is highlighted by the finding that Mer inhibition interacts synergistically in combination with multiple chemotherapeutic agents (methotrexate, vincristine, l-asparaginase, dexamethasone) to induce apoptosis. These chemotherapies are components of standard B-ALL regimens, both during initial therapy to decrease disease burden and during maintenance therapy to eradicate minimal residual disease. They mediate anti-leukemia activity by diverse mechanisms, suggesting that Mer inhibition may interact with a wide variety of cytotoxic agents. The increased apoptosis observed when Mer knockdown was combined with chemotherapy was accompanied by decreased activation of survival and proliferation signaling pathways, suggesting a biochemical mechanism by which Mer inhibition may impact leukemia cell survival. These data support the clinical application of Mer inhibition in combination with methotrexate and/or vincristine to increase therapeutic efficacy and potentially permitting dose reduction.
Even in the absence of a chemotherapeutic apoptotic stimulus, Mer knockdown B-ALL cell lines exhibited dramatically reduced colony formation in clonogenic assays, indicating that Mer inhibition alone is sufficient to impact growth and proliferation of B-ALL cells. Moreover, Mer knockdown using shRNA was sufficient to significantly delay onset and/or progression in an orthotopic mouse model of human leukemia, indicating an important role for Mer in leukemogenesis in vivo. Taken together, these results validate Mer as a therapeutic target in B-ALL, both alone and in combination with standard chemotherapy. Importantly, the experiments described in this manuscript demonstrating leukemogenic functions for Mer in B-ALL were performed by using two distinct cell lines transduced with two independent shRNA constructs, suggesting that this effect is specifically due to inhibition of Mer and not to an off-target effect of the shRNA.
Previous reports demonstrated that Mer prevents apoptosis by stimulating canonical cell survival pathways, including ERK1/2 and AKT.15,17,20 Here, we describe Mer-dependent modification of these signaling pathways and provide the first demonstration of p38 MAPK activation downstream of Mer in human B-ALL cells.
ERK1/2 has a well-defined role in protecting cancer cells from apoptosis. Activation of ERK1/2 has been associated with poor prognosis in ALL and a lower probability of achieving complete remission.31 In our study, inhibition of Mer expression prevented chemotherapy-induced activation of ERK1/2, indicating that aberrant expression of Mer enables B-ALL cells to activate the ERK1/2 survival pathway in response to cytotoxic agents. Similarly, AKT is known to promote cancer cell survival through prevention of apoptotic signaling.32 Activation of AKT in pediatric pre–B-ALL has been associated with poor blast clearance in induction and relapse,33 indicating a likely role in chemotherapy resistance. Previous studies have demonstrated that stimulation of Mer results in activation of AKT in murine leukemia cells and macrophages.17,27 Our findings validate these studies by showing that stimulation of Mer with the ligand Gas6 results in AKT activation in B-ALL cells, which is attenuated when Mer is inhibited. Similarly, when Mer is inhibited, signal transduction through AKT is reduced in B-ALL cells treated with methotrexate, suggesting a biochemical mechanism for the increased apoptosis observed.
While ERK1/2 and AKT are well-defined regulators of malignant cell proliferation and survival, the role of p38 is less clear. Activation of p38 has been associated with both pro- and antiapoptotic signaling in cancer models, depending on the cell lines used and the isoform of p38 that is activated.34 In this study, p38 phosphorylation was dependent on Mer in response to Gas6 stimulation, whereas phosphorylation of the related MAPK ERK1/2 was dependent on Mer in response to methotrexate. ERK1/2 and p38 share downstream signaling targets including MSK1/2 and the transcription factors CREB and ATF1; thus, Mer-dependent activation of these downstream signaling components may be dependent on the nature of the stimulus.
Although the role of Mer as an extracellular receptor and promoter of intracellular signal transduction has been previously reported, Mer has also been found to internalize and localize to the nucleus,35 suggesting that it may have a role in regulation of gene expression. Comparison of transcriptional differences between our control and Mer knockdown B-ALL cells after treatment with Gas6 or methotrexate demonstrated decreased prosurvival BCL-XL, PI3K, and PKC expression and increased proapoptotic BAX, NOXA, and PUMA expression in the knockdown cells, further demonstrating a survival advantage of lymphoblast signaling through Mer. Previous association between Mer inhibition and altered protein expression has been reported for BCL-XL,18 but the altered gene expression of PI3K, PKC, BAX, NOXA, and PUMA are novel findings not previously reported in the literature. The exact mechanism of Mer-related alterations in gene expression is not yet well understood and warrants further exploration.
The mechanism(s) of aberrant Mer expression in B-ALL and other cancers remains unknown. Given that E2A and PBX1 are both transcription factors, we considered the possibility that the E2A-PBX1 fusion protein drives expression of Mer in the t(1;19) subset of B-ALL. However, expression of E2A-PBX1 under control of an inducible promoter in an ALL cell line does not result in expression of Mer (K.K.S. and D.K.G., unpublished data). This result is consistent with our finding that Mer is expressed in both E2A-PBX1+ and E2A-PBX1− human B-ALL cell lines. Aberrant oncogene expression can occur via other mechanisms, including increased gene copy number, dysregulation of DNA methylation, or abnormal microRNA (miR) expression. Expression of the Mer-related receptor tyrosine kinase Axl has been inversely correlated with promoter methylation in acute myeloid leukemia cells.36 In metastatic breast cancer, overexpression of Mer correlated with loss of miR-335 and miR-126.37,38 Further studies are needed to assess whether these or other mechanisms contribute to ectopic expression of Mer in B-ALL.
In conclusion, these data demonstrate ectopic expression of Mer in pre–B-ALL. Inhibition of Mer expression reduces prosurvival signaling and clonogenic growth in methylcellulose, increases death of leukemia cells in response to treatment with cytotoxic chemotherapies, and significantly delays leukemogenesis in mouse models. These results validate Mer as a novel biologic target for the treatment of B-ALL. Although cure rates for B-ALL have considerably improved over the past 25 years, survivors remain at significant risk of late effects. Therapeutic agents that target Mer may directly inhibit leukemogenesis and enhance the efficacy of standard chemotherapy, potentially permitting dose reduction. While the focus of this study was pediatric leukemia, the widespread aberrant expression of Mer in human cancers suggests that novel agents that target Mer may also prove useful for treatment of adult leukemia and other Mer-expressing malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Stephen Hunger and the Children’s Oncology Group (grants CA98543 and CA114766) for providing E2A-PBX1+ samples and the University of Colorado Cancer Center Core Facilities (grant P30CA046934; flow cytometry, genomics and microarray, real-time PCR) for expert technical assistance.
This work was supported by grants from the American Academy of Pediatrics, the American Pediatric Society, and the National Institute of Child Health and Human Development (K12-HD000850) (A.B.L.-S. is a Fellow of their Pediatric Scientist Development Program); by the American Cancer Society (RSG-08-291-01-LIB) and National Institutes of Health (R01CA137078; D.K.G.), by Gabrielle’s Angel Foundation for Cancer Research (#030; D.K.G.), by the Damon Runyon Cancer Research Foundation (CI-39-07; D.K.G.), and by the Brent Eley Foundation (A.K.K.).
Authorship
Contribution: R.M.A.L., A.B.L.-S., D.D., R.A.C., K.M.J., A.M., L.N.B., A.W., K.K.S., A.K.K., and A.C.T. designed/performed experiments and analyzed data; X.L. and H.S.E. analyzed data; and R.M.A.L., A.B.L.-S., D.D., H.S.E., and D.K.G. wrote the manuscript.
Conflict-of-interest disclosure: D.K.G. and H.S.E. have filed patents on the targeting of the Mer tyrosine kinase as cancer therapy; and D.K.G., H.S.E., and D.D. have stock in Meryx, Inc (a company developing novel therapeutics against Mer). The remaining authors declare no competing financial interests.
Correspondence: Douglas K. Graham, Department of Pediatrics, P18-4401, Mail Stop 8302, Aurora, CO 80045; e-mail: doug.graham@ucdenver.edu.
References
Author notes
R.M.A.L. and A.B.L.-S. contributed equally to this work.

![Figure 5. Inhibition of Mer expression increases cell death and promotes apoptotic signaling pathway activation in response to chemotherapy. Apoptotic and dead cells were identified by flow cytometric analysis of cells stained with YO-PRO-1 iodide and PI. (A) Representative 697 flow cytometry profiles after treatment with methotrexate (MTX) are shown. The percentages of cells in early apoptosis (YO-PRO-1 positive, right lower quadrant), cells in later stages of apoptosis and necrosis (PI positive, upper quadrants), and live cells (left lower quadrant) are shown. Cultures of 697 and REH shControl, shMer1, and shMer4 cells were exposed to (B) 30 nM methotrexate, (C) 0.2 nM vincristine (VINC), (D) 50 nM (697) or 10 µm (REH) dexamethasone (DEX), or (E) 3 IU/mL l-asparaginase (L-ASP) or equivalent vehicle (phosphate-buffered saline [PBS] for methotrexate and vincristine; ethanol for dexamethasone and l-asparaginase) for 48 hours. Drug treated denoted as “+”, and vehicle treated denoted as “−”. Mean values and SEs derived from at least 3 independent experiments are shown. In response to methotrexate and vincristine, both REH and 697 Mer knockdown cells (shMer1 and shMer4) exhibited significant synergistic increases in the total percentage of apoptotic and dead cells compared to drug-treated shControl cells. In response to dexamethasone and l-asparaginase treatment, 697 cells exhibit significant induction of apoptosis. REH shMer cells exhibited a trend toward increased sensitivity in response to l-asparaginase, although there was no change in response to dexamethasone. *P < .05; **P < .01. (F) Whole-cell lysates were prepared from 697 and REH wild-type, shControl, shMer1, and shMer4 cells treated with methotrexate for 48 hours, and PARP, caspase 3, and caspase 8 were detected by immunoblot analysis. Representative 697 immunoblots are shown.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/9/10.1182_blood-2013-01-478156/4/m_1599f5.jpeg?Expires=1769083381&Signature=xN886FYzFAdOMyRkHqT0JAziBlkRA7XgJuR3FgLb~wqcPFfX10PWK8THjM3cl1HC4BO5qTdE0KLc9P2UphC~3UtRv2E-GLxzEKHJFUqW3aqTyvS6taw5Gtt3bkMLRW9KB3jKBlbS7d8-a-3~ni0EGWP4uZzJT4XLEn40Xum~Gi7eQcgGD3cJac832p1wOIp0jqQ8LpmSx9T74AP5aOD7bK61q2AzS1NURPAgeUXRCgNsSP~-KAlmH7CAAVVgYAthTSBhPEtdQH~lY0k23a54TLfXuiGh0ldLQ8zDdgyfomSiZ485zcpLnYqcl-IzNTsN1MkMwKkIZAoEa-s3JZM5Xg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)