Key Points
Complete genome sequence analysis of 40 DLBCL tumors and 13 cell lines reveals novel somatic point mutations, rearrangements, and fusions.
Recurrence of mutations in genes involved in B-cell homing were identified in germinal center B-cell DLBCLs.
Abstract
Diffuse large B-cell lymphoma (DLBCL) is a genetically heterogeneous cancer composed of at least 2 molecular subtypes that differ in gene expression and distribution of mutations. Recently, application of genome/exome sequencing and RNA-seq to DLBCL has revealed numerous genes that are recurrent targets of somatic point mutation in this disease. Here we provide a whole-genome-sequencing-based perspective of DLBCL mutational complexity by characterizing 40 de novo DLBCL cases and 13 DLBCL cell lines and combining these data with DNA copy number analysis and RNA-seq from an extended cohort of 96 cases. Our analysis identified widespread genomic rearrangements including evidence for chromothripsis as well as the presence of known and novel fusion transcripts. We uncovered new gene targets of recurrent somatic point mutations and genes that are targeted by focal somatic deletions in this disease. We highlight the recurrence of germinal center B-cell-restricted mutations affecting genes that encode the S1P receptor and 2 small GTPases (GNA13 and GNAI2) that together converge on regulation of B-cell homing. We further analyzed our data to approximate the relative temporal order in which some recurrent mutations were acquired and demonstrate that ongoing acquisition of mutations and intratumoral clonal heterogeneity are common features of DLBCL. This study further improves our understanding of the processes and pathways involved in lymphomagenesis, and some of the pathways mutated here may indicate new avenues for therapeutic intervention.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is an aggressive non-Hodgkin lymphoma (NHL) with at least 2 molecular subtypes that demonstrate distinct clinical outcomes and gene expression profiles. Because these cancers derive from mature B cells, the mutations that arise in DLBCLs can result from somatic hypermutation that targets a small number of genes,1 as well as structural rearrangements that arise from double-strand breaks that can be initiated by the B-cell recombination apparatus. In recent years, multiple groups have used massively parallel sequencing (genome/exome sequencing and RNA-seq) to ascertain the full set of genes targeted by somatic single-nucleotide variants (SNVs) in this disease.2-5 On the basis of these and earlier studies,6 it is now known that the 2 molecular subtypes also harbor distinct repertoires of somatic copy number alterations (CNAs) and SNVs. In particular, mutations affecting genes involved in B-cell receptor signaling and nuclear factor κB are common in the activated B-cell variety,7 whereas those affecting certain genes with roles in histone modification may be more common in the germinal center B-cell (GCB) subtype.2,8,9
These studies have confirmed that DLBCL is a genetically complex and heterogeneous disease and argue that a more complete understanding of the different mutation targets and mutational processes operating in this cancer is necessary. Previous studies have used mainly exome sequencing, which queries ∼1% of the genome, or RNA-seq to study DLBCL, but both methods are blind to the global patterns of mutation, as well as to the landscape of copy-neutral/balanced (or small) genomic rearrangements.2-5 To address this deficiency, our study reports the whole-genome sequencing (WGS) of 40 DLBCL tumor/normal pairs integrated with RNA-seq and copy number data from 96 tumors, and our analysis reveals a rich collection of SNVs, genomic rearrangements, and fusion transcripts. Overall, these data reveal new genes of interest as well as previously undescribed mutational modes operating in individual cases. We provide evidence for recurrent mutations affecting several of the novel genes in separate patient cohorts and the 13 DLBCL cell lines sequenced in this study. Our analysis infers the temporal ordering of mutations in the evolution of individual tumors and provides evidence that acquisition of driver mutations continues to occur during tumor progression, in addition to very early in tumorigenesis.
Materials and methods
This project was approved by the University of British Columbia–BC Cancer Agency Research Ethics Board as part of a broad effort to increase understanding of the molecular biologic characteristics of lymphoid cancers. Informed consent was obtained in accordance with the Declaration of Helsinki. Raw sequencing data are available by application through dbGAP (study accession: phs000532.v2.p1).
Sequencing
We sequenced tumor and matched constitutional DNA (peripheral blood) from 40 de novo DLBCL cases, using Illumina WGS to achieve between 27.9 and 56.6× average redundant coverage (median, 33.9×). These 40 cases are a subset of the 96 cases analyzed by copy number analysis (see below). To aid in determining the recurrence of mutations and identify suitable models for subsequent study of individual mutational events, we sequenced the genomes of 13 commonly studied DLBCL cell lines: DB, DOHH-2, Karpas422, NU-DUL-1, NU-DHL-1, OCI-Ly1, OCI-Ly3, OCI-Ly7, OCI-Ly19, SU-DHL-6, SU-DHL-9, MD903, and WSU-DLCL2. Libraries were sequenced on the Illumina HiSequation 2000 platform, according to Illumina protocols, generating paired-end 100-bp reads using a combination of v2 and v3 chemistry and HiSeq Control Software software versions 1.3.8 and 1.4.8.
SNV identification
SNVs were identified in genomes using SNVMix,10 as described.2 We further ranked the quality of candidate somatic calls using MutationSeq.11 On the basis of the verification results, we considered variants assigned a MutationSeq score of at least 0.2 to be high confidence, whereas for genome-wide mutation calling, to determine mutation spectrum and load, a threshold of 0.5 was used.
SNV verification and mutation recurrence testing
A subset of the somatic SNVs was confirmed using deep amplicon sequencing (supplemental Materials and methods, available on the Blood Web site). To determine the accuracy of our SNV identification approach, a mixture of high-confidence variants (5-10 per case) and variants with low MutationSeq probabilities were selected for verification (568 in total). Of the variants with sufficient coverage achieved, the verification rate of the entire set was 90.6%, and 96.2% for variants passing a MutationSeq score cutoff of 0.2.
For determining the recurrence of mutations in GNA13, each exon of this gene was amplified using polymerase chain reaction from 279 individual de novo DLBCL tumor samples. Eighty of the cases in this cohort were also previously analyzed by RNA-seq. Amplicons from individual patients were pooled, sheared by sonication, and constructed into indexed Illumina sequencing libraries, as previously described.12 Indexed libraries were pooled in batches of up to 92, and each pool was separately sequenced on a HiSeq2000 instrument using 100-bp reads, affording more than 100× coverage across all exons in most samples. These data were aligned to hg18 and analyzed for SNVs and indels, using SNVMix10 and SAMtools.13 Each candidate variant was manually inspected in an Integrative Genomics Viewer.14
Selective pressure analysis
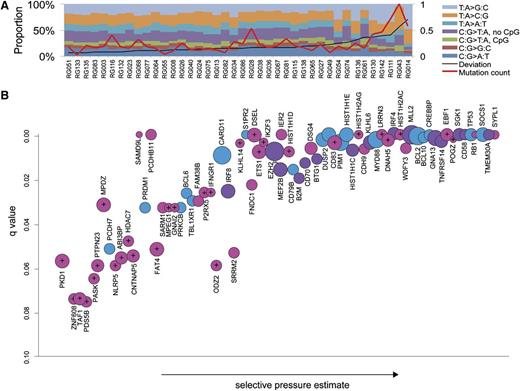
All high-confidence or experimentally verified silent and nonsilent SNVs identified in the 40 genomes were pooled. Selective pressure estimates were calculated, using the Greenman model as described,15 for any gene with 3 or more variants. We also included splice site mutations and separately estimated the selective pressure on this mutation type. The maximum of each of the 3 estimates was used to produce the gene order seen in Figure 1. Approximate P values were calculated by Monte Carlo simulation with 100 000 iterations, and these were adjusted using the Benjamini-Hochberg method (false discovery rate = 0.08).
Mutation spectra and significantly mutated genes. (A) The somatic point mutation spectrum observed genome-wide in each of the 40 cases. Overall, mutations affecting TA base pairs were more common than CG pairs, with TA>CG transitions the most common mutation observed on average. Some of the outliers, such as RG043, RG014, and RG111, harbored mutations in genes involved in DNA repair (supplemental Table 1; “Discussion”). (B) Mutated genes with significant evidence for positive selection (false discovery rate = 0.08) are ordered on the x-axis based on selective pressure estimate. The y-axis shows the adjusted P value such that highly significant genes, typically because of a larger number of observed mutations, lie toward the upper right. The size of the circles is proportional to the number of cases in the patient cohort in which a nonsilent or splice site SNV was identified. Significant genes identified in the larger patient cohort in our previous RNA-seq study are purple, and those identified in separate studies3-5 are blue. The remaining 41 genes shown in pink have not, to our knowledge, been identified by others as significant targets of point mutation in DLBCL. Genes denoted with crosshairs indicate those with secondary support for mutations from other studies or the 13 DLBCL cell lines sequenced here (see supplemental Table 2 for details and references). Genes affected by splice site mutations included the known tumor suppressor genes MLL2, RB1, CREBBP, and TP53, as well as others with signatures indicative of inactivation, including DNAH5 and SGK1.
Mutation spectra and significantly mutated genes. (A) The somatic point mutation spectrum observed genome-wide in each of the 40 cases. Overall, mutations affecting TA base pairs were more common than CG pairs, with TA>CG transitions the most common mutation observed on average. Some of the outliers, such as RG043, RG014, and RG111, harbored mutations in genes involved in DNA repair (supplemental Table 1; “Discussion”). (B) Mutated genes with significant evidence for positive selection (false discovery rate = 0.08) are ordered on the x-axis based on selective pressure estimate. The y-axis shows the adjusted P value such that highly significant genes, typically because of a larger number of observed mutations, lie toward the upper right. The size of the circles is proportional to the number of cases in the patient cohort in which a nonsilent or splice site SNV was identified. Significant genes identified in the larger patient cohort in our previous RNA-seq study are purple, and those identified in separate studies3-5 are blue. The remaining 41 genes shown in pink have not, to our knowledge, been identified by others as significant targets of point mutation in DLBCL. Genes denoted with crosshairs indicate those with secondary support for mutations from other studies or the 13 DLBCL cell lines sequenced here (see supplemental Table 2 for details and references). Genes affected by splice site mutations included the known tumor suppressor genes MLL2, RB1, CREBBP, and TP53, as well as others with signatures indicative of inactivation, including DNAH5 and SGK1.
Mutation spectrum determination
Mutation spectra for each case were computed by summing the 7 distinct mutation types for genome-wide somatic mutation calls (CG>AT, CG>GC, C*G>TA, CG>TA, TA>AT, TA>CG, and TA>GC, with C* indicating a CpG context cytosine in the reference genome). Proportional mutation spectra were computed by normalizing total mutations to 1, and average proportional spectrum across all samples was determined by taking the mean of the proportions. Spectrum deviation was computed as the sum of the differences between the proportions of each mutation type in a sample vs the average.
Genomic rearrangement and fusion transcript discovery by de novo assembly
RNA-seq libraries from the patient samples and cell lines were assembled using ABySS (version 1.2.5) and the empirically-determined k-mer values k26-k50, as described.16 Tumor genomes were assembled with version 1.2.6 of ABySS, using a crucible assembly (supplemental Materials and methods). RNA-seq contigs supporting the presence of a fusion transcript were further annotated for their effect on the affected genes. We attempted verification for each fusion event and a subset of rearrangements (71) identified by WGS (supplemental Materials and methods).
Integrative analysis of all mutation types
The 96 DLBCL cases were analyzed for somatic CNAs, using Affymetrix SNP6.0 data (supplemental Materials and methods). CNA information derived from the WGS data (40 cases) and array-derived CNAs from the additional 56 cases were used for this analysis. To identify genes recurrently mutated in DLBCL, we counted the number of cases in which each gene is affected by any focal mutation, including somatic nonsilent SNVs, small (< 100 nt) somatic indels, small deletions/CNAs (< 50 kb), and chromosomal breakpoints of other rearrangements. For the breakpoints of other structural rearrangements, any gene within 250 kb of either breakpoint was considered potentially relevant. When the relevant gene was known (eg, BCL2, BCL6), other genes near that breakpoint were not considered. To integrate these different sources of data to collectively identify potential “driver” genes, a probabilistic model (DriverNet)17 was applied. This method incorporates the above data types and pathway information to determine the mutations that are likely to result in perturbations to gene expression.
Duplication and mutation timing
Estimates of the relative time at which duplication events arose relative to somatic SNVs (“timings”) were computed using the approach described by Nik-Zainal et al,18 based on computation of mutation ploidy by Greenman et al.19 Calculation was based on integer copy number data from cnaseq,20 loss of heterozygosit (LOH) and allelic ratio data from APOLLOH,21 pathology-derived normal contamination estimates, genome-wide SNV calls from SNVMix10 and MutationSeq,11 and allele count information derived using SAMtools.13 Point estimates for the proportional timing of all duplications in the DLBCL genomes were inferred from these data (supplemental Materials and methods). For each timed segment, 95% confidence intervals were computed, using a bootstrapping approach with 10 000 iterations of resampling the original mutations and recomputing timings. Copy number segments smaller than 100 kb were excluded from analysis. Duplications involving multiple sequential events cannot be timed precisely and were approximated. Adjacent copy number segments were used to infer timing of events in samples in which focal amplification is accompanied by larger events.
Estimating cellular frequency/clonality of SNVs
Clonality estimates were derived by integrating deep amplicon sequencing data from the SNV validation experiment and both CNA and LOH data from the 40 cases analyzed by WGS. Using these data, PyClone version 0.9.0 was run, using 10 000 iterations of the MCMC chain; the first 1000 samples were discarded as burn-in, and the remaining MCMC samples were used to estimate the posterior distribution on cellular frequencies, using kernel density estimation. Priors for PyClone were set by giving equal prior probability to all genotypes containing at least a single mutant allele that was compatible with the predicted copy number. Equal prior weight was also given to states in which cancer cells lacking the mutation were predicted to have copy number 2 or the copy number of the mutated cells. Tumor content estimates, derived from taking the minimum of the computational and pathology predictions, were input to PyClone (http://compbio.bccrc.ca/software/pyclone/).
Results
Mutation patterns and significantly mutated genes
Unlike RNA-seq or exome sequencing, WGS provides the opportunity to globally determine the pattern, frequency, and location of somatic point mutations across the entire tumor genome. We observed nonrandom patterns of SNV distribution across the genomes with particular enrichment near transcription start sites, an observation that is consistent with aberrant somatic hypermutation (aSHM; catalyzed by AID), which we have described elsewhere.22 Beyond this local variability in mutation rates, we also detected distinct mutational patterns and sequence contexts across individual patients. There was a broad range of overall somatic mutation load among the sequenced genomes, with the total number of candidate somatic SNVs detected per case ranging from 1165 to 48 385. We determined that the case with the lowest number of SNVs contained significant contamination from normal cells, and after excluding this case, we found an average of 12 086 somatic mutations (4.21 mutations/Megabase genome-wide) and 205.6 nonsilent mutations per genome (range, 35-400; supplemental Table 1). Despite the variability in mutation load, the spectrum of SNV types and sequence contexts was largely consistent across cases, with some notable exceptions (Figure 1A; supplemental Figure 1; “Discussion”).
Our previous RNA-seq-based mutation screen2 was insensitive to splice site mutations and was biased toward detecting SNVs in genes that were actively transcribed in DLBCL, thus potentially restricting our ability to discover tumor suppressor genes. Despite the smaller cohort analyzed here, our selective pressure analysis of the SNVs detected in these 40 genomes identified 74 significant genes, including many of those reported in our previous study2 or subsequent exome-based studies,3-5 while also capturing 41 genes not previously reported as significantly mutated in DLBCL (Figure 1B). To inform on the potential recurrence of mutations in any of the 74 genes in other data sets, we analyzed 13 DLBCL cell lines by WGS and also mined the full data sets from other large DLBCL patient cohorts (supplemental Table 2).3-5
The pattern of mutations within individual genes can provide additional insight into the potential function of a gene as an oncogene or tumor suppressor and can also indicate the result of aSHM. Eleven of the newly detected genes had mutation signatures indicative of tumor suppressor function with mutations at splice sites (eg, EBF1, RB1) or producing a truncated protein (eg, DNAH5). Some of these had low sequence coverage in our previously analyzed RNA-seq libraries (supplemental Table 2), whereas others had mutated splice sites. Mutation hot spots within a gene (eg, recurrently affecting a codon) are indicative of potential oncogenes, and these were observed in a subset of the genes detected. In the genes with signatures of aSHM (IRF4, IRF8, BCL6, PIM1, CD83, P2RY8, BCL2, and DUSP2), hot spots may reflect preference of AID for certain sequence motifs rather than evidence of selection. The remaining genes with recurrent mutations and patterns inconsistent with SHM include those with known dominant acting mutations in lymphoma (MYD88, CARD11, CD79B, and EZH2) as well as TBL1XR1, MEF2B, FAT4, PKD1, NLRP5, and DSEL. Hot spots in MEF2B2 and TBL1XR14,23 have been reported in multiple NHL types, but the function of mutations in these or the remaining genes has not been elucidated.
A separate set of genes harbored an excess of missense mutations but no observable hot spots. One notable observation was the mutation of multiple genes that encode histone proteins, an observation that has previously been reported in DLBCL and other cancers.4,24 Seventeen of the genomes had at least a single nonsilent SNV affecting a gene encoding either linker histone protein H1 or core protein H2 (supplemental Figure 2). The recurrence of mutations in these genes was confirmed in many of the DLBCL cell lines (supplemental Table 3), but the potential function of these mutations remains unclear. Other recurrent mutation targets included cell surface receptors mitigating interactions with T cells, such as CD7025 and CD8326 ; purinergic receptors (P2RY8 and P2RX5), as well as cytokine receptors (S1PR2); and Gα subunits of G-protein-coupled receptors (GNAI2).
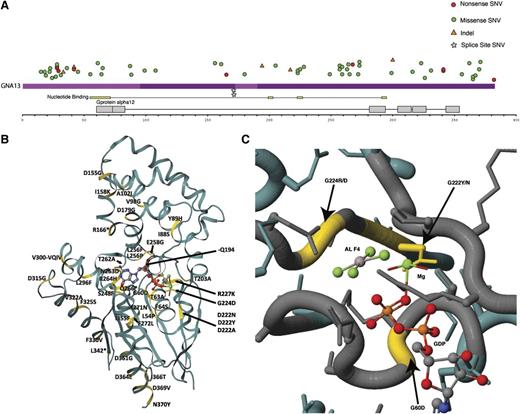
We previously identified GNA13, another Gα protein, as a recurrent target of inactivating mutations in DLBCL2 and noted that S1PR2 is a target of aSHM.22,27 As S1P2 (encoded by S1PR2) can couple to each of these Gα proteins to regulate B-cell migration and homing (supplemental Figure 3),28,29 we further explored the mutation patterns in each gene (Figure 230 ; supplemental Figure 4 and supplemental Table 4). This analysis clarified the pattern that mutations affecting GNA13 were typically inactivating and confirmed that these mutations are strongly enriched in GCB cases (P = 1.289 × 10−8, Fisher’s exact test). Using approaches described in our recent study,12 we also tested whether the presence of GNA13 mutations was prognostic in the 178 cases uniformly treated with R-CHOP but found no significant correlation with patient outcome (supplemental Figure 5).
Mutations affecting GNA13 in a large cohort of DLBCLs. Guided by the prevalence of GNA13 mutations in our DLBCL cohorts analyzed by RNA-seq and WGS, we sought to ascertain the full mutational landscape of this gene across a large number of de novo DLBCL cases (n = 279), of which 182 had been classified as GCB or non-GCB using immunohistochemistry.30 (A) Nonsilent SNVs, indels, or splice site mutations were detected in a total of 40 patients (14.3%), with many cases harboring more than a single mutation. Up to five nonsilent mutations affecting GNA13 were observed in one patient. Overall, multiple truncation inducing mutations including frameshift indels and introduced stop codons were observed. The ratio of transitions to transversions and the large number of mutations affecting the WRCY/RGYW motif is consistent with AID-mediated mutation; however, there was no observable enrichment of mutations in the 5ʹ end of the locus (supplemental Table 5). In agreement with our previous observation, GNA13 mutations were strongly enriched in GCB, with 29 of 89 GCB (32.6%) cases having at least a single mutation in this gene and only 2 of 91 non-GCB cases mutated. (B) We mapped each of the mutations to the solved structure of Galpha13 (PDB accession no. 3AB3) and observed some nonsynonymous mutations in close proximity to the catalytic site (C), including multiple residues that interact directly with the substrate (GTP). Taken in conjunction with the prevalence of truncating mutations, we predict these likely inhibit the signaling activity of Gα13.
Mutations affecting GNA13 in a large cohort of DLBCLs. Guided by the prevalence of GNA13 mutations in our DLBCL cohorts analyzed by RNA-seq and WGS, we sought to ascertain the full mutational landscape of this gene across a large number of de novo DLBCL cases (n = 279), of which 182 had been classified as GCB or non-GCB using immunohistochemistry.30 (A) Nonsilent SNVs, indels, or splice site mutations were detected in a total of 40 patients (14.3%), with many cases harboring more than a single mutation. Up to five nonsilent mutations affecting GNA13 were observed in one patient. Overall, multiple truncation inducing mutations including frameshift indels and introduced stop codons were observed. The ratio of transitions to transversions and the large number of mutations affecting the WRCY/RGYW motif is consistent with AID-mediated mutation; however, there was no observable enrichment of mutations in the 5ʹ end of the locus (supplemental Table 5). In agreement with our previous observation, GNA13 mutations were strongly enriched in GCB, with 29 of 89 GCB (32.6%) cases having at least a single mutation in this gene and only 2 of 91 non-GCB cases mutated. (B) We mapped each of the mutations to the solved structure of Galpha13 (PDB accession no. 3AB3) and observed some nonsynonymous mutations in close proximity to the catalytic site (C), including multiple residues that interact directly with the substrate (GTP). Taken in conjunction with the prevalence of truncating mutations, we predict these likely inhibit the signaling activity of Gα13.
Structural alterations and fusion transcripts
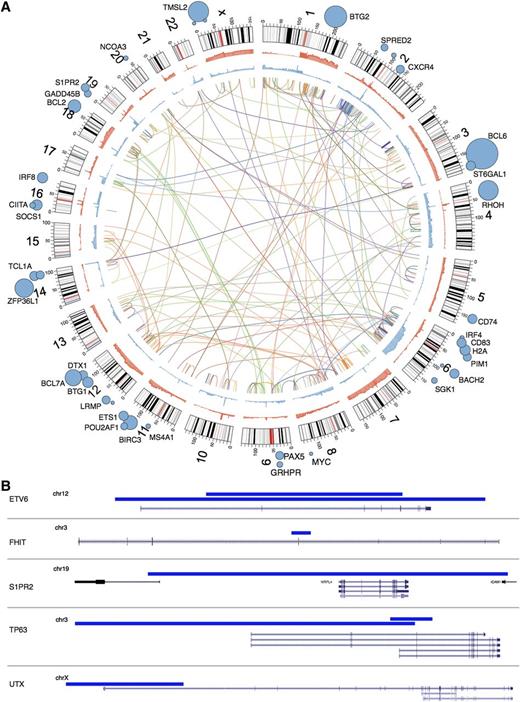
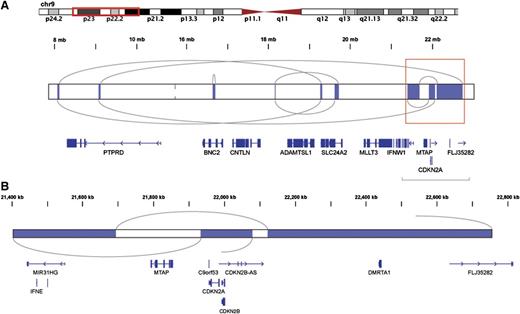
Our de novo assembly-based approach allowed us to discover genomic rearrangements and resolve their breakpoint sequences (Figure 3A31 ; “Materials and methods”). Coverage-based DNA copy number analysis showed that many of these breaks were associated with changes in copy number state and also indicated copy-neutral events, including inversions and translocations. The number of individual events detected in a genome ranged from zero to 41 (supplemental Tables 1 and 5). The pattern of rearrangements and CNAs in some of the highly rearranged genomes is consistent with chromothripsis (Figure 4).32 Separate cases included evidence for complex rearrangements resulting in high-level amplification of the REL locus via a distinct mechanism (supplemental Figure 6 and supplemental Figure 7). Some of the deletions detected by assembly were below the detection resolution of CNA approaches (Figure 3B). Unlike the large CNAs, which typically involve entire chromosomes or chromosomal arms (supplemental Figure 8), these commonly affected single genes or portions thereof, allowing their functional effect to be more readily predicted.
Overview of rearrangements, CNAs, SNVs, SHM and focal deletions detected. (A) Inner arcs represent somatic rearrangements from each of the patient genomes, with a different color depicting each case. Cumulative summaries of all the somatic CNAs detected across all 96 cases are depicted in blue (deleted regions) and red (amplified regions). SHM targets identified from these genomes22 are indicated with blue circles with diameter proportional to the number of mutated cases. (B) Small deletions are often not detectable by copy number analysis methods. Our de novo assembly-based pipeline identified breakpoints representing small deletions (indicated by blue bars), some of which affected a single gene. Two cases were found to have such deletions affecting ETV6. Of note, a fusion involving ETV6 and the immunoglobulin heavy chain locus was observed in a separate case. Deletions affecting other genes likely to be relevant to DLBCL are also shown. FHIT, with a focal deletion shown here, was also a common target of larger deletions by CNA analysis. S1PR2 was also a significant target of somatic point mutations and functionally cooperates with proteins encoded by GNA13 and GNAI2 (“Discussion”). The 2 deletions affecting TP63 in a single case are also shown (see supplemental Figure 12). The upper 3 transcripts represent TA isoforms, whereas the lower 2 correspond to Δ N isoforms. UTX is a histone demethylase that acts on H3K27, the same lysine targeted by EZH2, which is a target of activating mutations in NHL. A recently described small molecule inhibitor of EZH2 activity showed efficacy in DLBCL cell lines with UTX mutations.31
Overview of rearrangements, CNAs, SNVs, SHM and focal deletions detected. (A) Inner arcs represent somatic rearrangements from each of the patient genomes, with a different color depicting each case. Cumulative summaries of all the somatic CNAs detected across all 96 cases are depicted in blue (deleted regions) and red (amplified regions). SHM targets identified from these genomes22 are indicated with blue circles with diameter proportional to the number of mutated cases. (B) Small deletions are often not detectable by copy number analysis methods. Our de novo assembly-based pipeline identified breakpoints representing small deletions (indicated by blue bars), some of which affected a single gene. Two cases were found to have such deletions affecting ETV6. Of note, a fusion involving ETV6 and the immunoglobulin heavy chain locus was observed in a separate case. Deletions affecting other genes likely to be relevant to DLBCL are also shown. FHIT, with a focal deletion shown here, was also a common target of larger deletions by CNA analysis. S1PR2 was also a significant target of somatic point mutations and functionally cooperates with proteins encoded by GNA13 and GNAI2 (“Discussion”). The 2 deletions affecting TP63 in a single case are also shown (see supplemental Figure 12). The upper 3 transcripts represent TA isoforms, whereas the lower 2 correspond to Δ N isoforms. UTX is a histone demethylase that acts on H3K27, the same lysine targeted by EZH2, which is a target of activating mutations in NHL. A recently described small molecule inhibitor of EZH2 activity showed efficacy in DLBCL cell lines with UTX mutations.31
A likely chromothripsis event resulting in loss of the CDKN2A/B locus. Shown are regions of somatic copy number loss (blue) detected by HMMCopy analysis of a single case. Gray arcs represent rearrangement breakpoints and connections, as determined by contigs resulting from whole genome assembly of that case. (A) The rearranged region includes a series of deleted segments and encompasses many genes. The coordinated loss of genetic material and focused rejoining of fragments in these discrete regions is indicative of a single mutational event followed by DNA repair in a single cell cycle and is consistent with the chromothripsis model.32 (B) An expanded view of the boxed region from (A) is shown. One of the deleted segments encompasses both CDKN2A and CDKN2B, known to be targets of focal deletion in DLBCL6 and also found to be commonly deleted in this cohort.
A likely chromothripsis event resulting in loss of the CDKN2A/B locus. Shown are regions of somatic copy number loss (blue) detected by HMMCopy analysis of a single case. Gray arcs represent rearrangement breakpoints and connections, as determined by contigs resulting from whole genome assembly of that case. (A) The rearranged region includes a series of deleted segments and encompasses many genes. The coordinated loss of genetic material and focused rejoining of fragments in these discrete regions is indicative of a single mutational event followed by DNA repair in a single cell cycle and is consistent with the chromothripsis model.32 (B) An expanded view of the boxed region from (A) is shown. One of the deleted segments encompasses both CDKN2A and CDKN2B, known to be targets of focal deletion in DLBCL6 and also found to be commonly deleted in this cohort.
Analysis of genomic sequence along with RNA-seq data can identify the fusion products that can result from rearrangements. In the entire cohort (96 patients), we detected 130 fusion transcripts among 64 cases, but no novel recurrent fusions beyond those involving TP63, which have been described elsewhere33 (supplemental Figures 9 and 10; supplemental Table 6). The remaining fusion events resulting in preserved reading frames, along with validation data, are provided in supplemental Figure 11. Two other fusions involving TP63 (with GAS2 and TMEM110) were also observed, but neither contained a fused open reading frame. The case harboring a GAS2 fusion was observed in a sample also containing a TP63-TBL1XR1 fusion. The second fusion joined the 3′ UTR of GAS2 to the second exon of TP63. The case involving TMEM110 included the 5′ UTR of TP63 and the bulk of TMEM110. We compared the expression of TP63 in cases lacking any fusion with all cases with fusions affecting TP63 and found that the presence of a fusion correlated with increased expression of TP63 transcript (RPKM; P = .00378, Wilcoxon rank sum test). The elevated abundance of TP63 transcripts in cases with this fusion has been previously confirmed by quantitative reverse transcription-polymerase chain reaction.33 In one case, we identified 2 separate deletions affecting TP63. The larger deletion (∼363 kb) removed the first 3 exons that encode the larger TA-TP63 isoforms, whereas the smaller deletion (45 kb) removed the first exon of the shorter ΔN isoform, as well as the fourth exon, which is shared by both isoforms (Figure 3B; supplemental Figure 12). The case harboring these deletions and 3 of the cases with fusions had evidence for monoallelic expression of TP63. Taken together with the elevated abundance of TP63 mRNA, this supports the notion that fusion with TBL1XR1 increases the abundance of TP63 message.
Integrative analysis of distinct mutational modes and gene expression
Although some genes tended to be mutated by a single process, such as SNVs or small indels (eg, EZH2 and MLL2) or rearrangement/deletion (eg, TP63), others were affected by a diverse combination of mutation types. To provide a more complete view of the genes affected by various mutation types, we determined the number of times each gene was mutated in any modality (excluding large deletions; supplemental Tables 7-9). The genes most commonly altered include well-studied lymphoma-related genes such as CDKN2A/B, BCL2, and BCL6. Supplemental Figure 13 gives an overview of the prevalence and distribution of various mutation types affecting these and some of the additional commonly altered genes in the large cohort. Novel genes highly ranked by this approach include CDK11A, which was commonly lost by focal deletions or large deletions encompassing 1p36 (supplemental Figure 14); P2RY8 and CSMD3, each with a combination of SNVs and focal deletions; and TBL1XR1. The involvement of TBL1XR1 in fusions with TP63,33 focal deletions,3 and SNVs4 in DLBCL has been reported previously, and our data further support these findings. As a complementary approach, we performed an analysis that integrates the mutation data (CNAs, SNVs, and indels) and gene expression information from the RNA-seq data to determine whether mutations result in perturbations to the gene expression network in trans (“Materials and methods”). This analysis indicated that mutations in many of the 74 genes identified as significantly under selection or commonly affected by other alterations might perturb gene expression in DLBCL (supplemental Figure 15).
Dissecting temporal acquisition of mutations and clonal substructure
Determining the order in which individual somatic alterations arise can provide insight into the roles of individual genes and mutations in oncogenesis. One such method to approximate the order in which mutations arise involves comparing the clonal frequency of SNVs within a tumor. We and others have used this to determine the clonal (early) and subclonal (late) mutations in breast cancer34 and leukemia.35 By deeply resequencing a subset of the somatic SNVs detected in these genomes and correcting allelic counts for CNAs and LOH, we produced estimates of the cellular frequency of each of these mutations (“Materials and methods”). Examples of these estimates for 3 tumors are shown in supplemental Figure 16. A commonly held assumption is that important driver mutations are acquired early in cancer development, and thus would be present in all cells of the tumor.36 Surprisingly, and counter to this, we observed multiple examples of well-characterized driver mutations, including hot spot mutations in EZH2, MYD88, CARD11, and CD79B, that were present in subclonal populations.
Using WGS data, it is also possible to approximate the temporal ordering of copy number gains.18 In cases in which both SNVs and large duplications were present in the same tumor, we inferred the order in which these occurred (“Materials and methods;” supplemental Materials and methods).18,19,37 We first focused on gains of the REL locus, as these are among the only common focal amplifications in DLCBL. We found that, similar to the known driver point mutations, REL amplifications can arise both early and late in tumor evolution (supplemental Figure 17). We next approximated the timing of all amplified regions in each of the 40 genomes (supplemental Table 10). This analysis showed that duplications affecting chromosomes 18 (targeting BCL2) and 6p often occurred quite early in cancer development, whereas duplications of chromosomes 7 and 21 were among the latest events (supplemental Figure 18). Despite the trend toward earlier acquisition, we found examples of +18 (BCL2) and +8q (MYC) arising as late events in some tumors (Figure 5).38 Other regions of amplification that were commonly observed are also shown for comparison, along with candidate oncogenes indicated in each region.
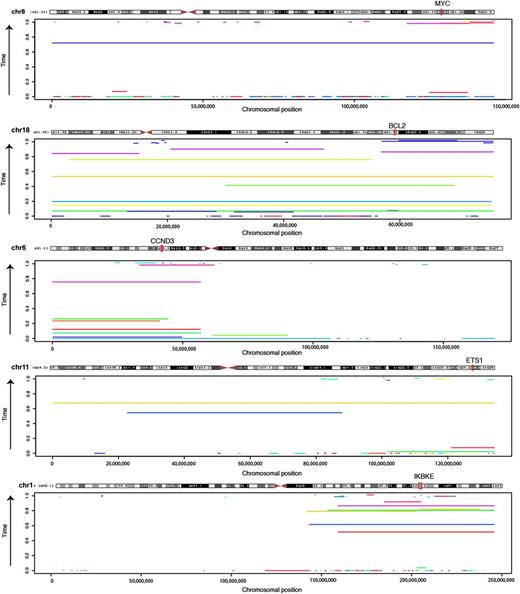
Timing of chromosomal duplications in DLBCL evolution. The sequence data can be used to approximate the relative time in which individual large amplifications/gains occurred during the evolution of the tumor. Here, the timing estimate for amplifications detected in each genome is shown for 5 chromosomes commonly affected by such events. The genomic coordinates amplifications are shown on the x-axis, with separate colors indicating events detected in different individuals. The y-axis shows the time at which the event was estimated to occur, with events near the bottom arising earlier in tumor development and those near the top arising later. Only events for which we could precisely calculate timing were included (confidence interval range < 0.2). Samples involving approximated REL amplifications are shown separately (supplemental Figure 17). Despite arising later in some cases, gains of 18 were nonetheless one of the earliest of all amplification events detected (supplemental Figure 18). The genes targeted by amplifications of 11q and 1q have not been conclusively identified. The position of ETS1 is indicated because it also a significant target of somatic point mutations in our data, and thus a potential novel oncogene. The region of overlap between the regions gained on 1q contains a small number of genes including IKBKE, which encodes IkappaB kinase ε, a positive regulator of RELA.38 Among the 96 DLBCL cases analyzed for CNAs, IKBKE expression was significantly higher in amplified cases (P = .00745, Wilcoxon Rank Sum test).
Timing of chromosomal duplications in DLBCL evolution. The sequence data can be used to approximate the relative time in which individual large amplifications/gains occurred during the evolution of the tumor. Here, the timing estimate for amplifications detected in each genome is shown for 5 chromosomes commonly affected by such events. The genomic coordinates amplifications are shown on the x-axis, with separate colors indicating events detected in different individuals. The y-axis shows the time at which the event was estimated to occur, with events near the bottom arising earlier in tumor development and those near the top arising later. Only events for which we could precisely calculate timing were included (confidence interval range < 0.2). Samples involving approximated REL amplifications are shown separately (supplemental Figure 17). Despite arising later in some cases, gains of 18 were nonetheless one of the earliest of all amplification events detected (supplemental Figure 18). The genes targeted by amplifications of 11q and 1q have not been conclusively identified. The position of ETS1 is indicated because it also a significant target of somatic point mutations in our data, and thus a potential novel oncogene. The region of overlap between the regions gained on 1q contains a small number of genes including IKBKE, which encodes IkappaB kinase ε, a positive regulator of RELA.38 Among the 96 DLBCL cases analyzed for CNAs, IKBKE expression was significantly higher in amplified cases (P = .00745, Wilcoxon Rank Sum test).
Discussion
Previous publications exploring genomic sequences of DLBCL tumors focused only on the exonic portion of these tumors and the protein-altering SNVs. Together, the analyses presented here demonstrate, for the first time, the complete landscape of the diverse somatic mutation types that arise in a large number of DLBCL genomes. These data provide a global view of the variable mutation load and spectra that arise in DLBCL and further support the widely appreciated view of DLBCL as a genetically heterogeneous disease. Overall, these data indicate that the average mutation load in DLBCL is well above the level previously determined using exome sequencing in a small cohort.3 There were clear outliers in this cohort with respect to mutation load and the spectrum of mutation types. For example, RG043 had nearly twice the proportion of TA>CG mutations as the average tumor genome. This case also had the greatest mutation load overall, reaching almost 5 times the average across these genomes. Among these mutations, we found a single base indel in MSH3, which would result in a truncated protein. The association between MSH3 inactivation and increased mutation rates has been described in colorectal cancer.39 We noted that some of the other individual cases with distinct mutation spectra harbored mutations in genes encoding DNA polymerases (eg, POLE), but this observation was not consistent for all outliers. Overall, there was a substantial positive correlation indicating that samples with higher numbers of mutations also have more distinct mutation spectra (r = 0.69). Taken together, this suggests that unique mutational processes, resulting in an overall greater and distinct mutation load, are active in a subset of these tumors.
Despite the small cohort size, our study has uncovered additional recurrent targets of somatic SNVs. With the addition of GNAI2, somatic mutations affecting each of 3 separate genes that cooperate in Rho-mediated B-cell homing now been identified (GNAI2, S1PR2, and GNA13; supplemental Figures 3 and 4). We previously reported the recurrence of inactivating mutations in GNA13 and extend that observation here by determining this gene to be mutated in approximately 20% of de novo DLBCLs and up to 33% of GCB cases. Although not yet confirmed in a larger cohort, mutations affecting GNAI2 and S1PR2 (including the deletion shown in Figure 3) were restricted to GCB cases in our cohort of 96 patients. Interestingly, GNAI2 had a mutation pattern distinct from GNA13, with an apparent enrichment for missense mutations rather than truncating mutations. Some of the residues we found mutated in GNAI2 are orthologous to gain-of-function mutations that have been characterized in GNAI3 (supplemental Figure 4), and thus could be comparable to the oncogenic GNAS mutations common among pituitary tumors40 and pancreatic adenocarcinoma.41
Given the potential for activating mutations in GNAI2, we predict that each of the mutations observed in these 3 genes has an equivalent effect on the signaling events downstream of S1P2. In B cells, S1P concentration gradients are detected by S1P2 and serve to restrict cells to germinal centers.42 Signaling of S1P2 through Gi2 and Gα13 has distinct effects on Rho and Rac activity, and each protein has an opposing effect on PI(3)K/Akt signaling, with Gα13 leading to suppressed Akt activity and Gi2 promoting Akt.43 Recently, recurrent mutations in RHOA have also been observed in Burkitt lymphoma,44 which is another cancer known to commonly harbor GNA13 mutations.45 Although RHOA was not significantly mutated in this study, we note the presence of a single somatic mutation in our patient cohort (R68P) and a second mutation in the cell line OCI-Ly19 (Y66N). Together, these data further affirm an oncogenic role of perturbations to Rho-mediated B-cell migration or the indirect effect these pathways have on PI(3)K/Akt signaling, particularly within the GCB subgroup of DLBCL.
Beyond SNVs, our integrative analyses highlight the many genes and pathways mutated by other mechanisms, including many known to be important for DLBCL such as modulators of cell cycle and both p53 and Rb signaling (supplemental Figure 15). Although deletions affecting CDKN2A/B, RB1, and TP53 have all been described,46 splice site mutations affecting RB1, which we observed at high prevalence (7.5%), are a novel observation. Similarly, loss of CDKN2A by chromothripsis has not, to our knowledge, been described. In addition to these, we have identified TAF1 as a target of both nonsilent somatic SNVs and high-level amplifications (Figures 1; supplemental Figure 5). TAF1 encodes a transcription factor with both histone acetyltransferase and kinase activities, and the latter is regulated directly by Rb.47 In addition to its role as a general transcription factor, TAF1 regulates p53 activity by phosphorylating it at Thr55, thereby inducing its cytoplasmic shuttling and MDM2-mediated degradation.48,49 Given the prevalence of somatic events affecting both RB1 and TAF1 in this cohort, the role of these mutations in p53 signaling and potential avenues for therapeutic intervention should be further investigated.
Finally, these data provided an unprecedented opportunity to explore the relative temporal order of mutation acquisition in tumors. We explored the focal amplification of the REL locus in detail. Although REL amplification appeared to commonly arise early in cancer development, we also found evidence for continued increases in REL copy number over protracted time and even observed REL amplification quite late in some cases. In contrast, amplifications encompassing the BCL2 locus were more typically acquired early in tumorigenesis, in line with the widely held view that BCL2 deregulation is an early event in transformation. Nonetheless, we also provide counterexamples in which amplification of BCL2 and MYC arose later in tumor development, supporting that these genes can contribute to tumor progression even if not acquired early. By studying the allele frequency of individual SNVs in our validation cohort, we could also estimate the relative proportion of cells harboring certain mutations in each tumor. That analysis indicates, in accordance with a recent study of FL,36 that DLBCLs also undergo multiple rounds of clonal expansion. Interestingly, in contrast to the conclusion made in that study, our data indicate that important driver mutations including those in TP53, CARD11, MYD88, and CD79B can all be acquired during later steps in this process (supplemental Figure 16).
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the expert technical assistance from staff at the Genome Sciences Centre and project management from Dr Karen Novik. The authors gratefully acknowledge Dr Daniela Gerhard for helpful discussions.
The authors acknowledge the support of the BC Cancer Foundation, the Cancer Research Society, Genome Canada, and Genome BC. This project has been funded in whole or in part with federal funds from the National Institutes of Health National Cancer Institute (NO1-CO-12400). The authors also gratefully acknowledge funding support from the Cancer Research Society. This research was funded in part by The Terry Fox Research Institute (grant 019001). R.D.M. is supported by a British Columbia Cancer Foundation Investigator Establishment Award and M.A.M. is the UBC Canada Research Chair in Genome Sciences.
Authorship
Contribution: R.D.M. produced Figures 1 through 5 and, with E.P. and M.A.M., wrote the manuscript; E.P., A.R., and S.S. performed the mutation timing and clonal frequency analyses; J.D. performed DriverNet analysis and produced supplemental Figure 15; R.G. performed selective pressure analysis; A.J.M., R.C., I.B., and K.M. identified and annotated fusions and genomic rearrangements; R.D.C., N.F., F.C.C., and S.R. performed copy number analysis on the WGS and SNP6 data; M.M.-L., D.L.T., M.B.-M., R.D.H., and G.T. experimentally validated mutations and fusion events; A.H.K. performed hypermutation analysis; J.P. analyzed mutations in genes encoding core histone proteins; E.L.L. analyzed miRNA-seq data; R.M. and R.H. coordinated the sequencing; D.W.S., N.A.J., S.B.-N., B.M., and B.W. prepared the samples and performed fluorescence in situ hybridization, and D.W.S. performed the survival analysis for GNA13; and S.J., C.S., J.M.C., R.D.G., and M.A.M. conceived of the study, directed the analysis, and contributed to the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marco A. Marra, Genome Sciences Centre. 570 West 7th Ave, Vancouver, BC, Canada; email: mmarra@bcgsc.ca.