In this issue of Blood, Krawitz et al report on a patient with paroxysmal nocturnal hemoglobinuria (PNH) who does not have a mutation of PIG-A, but in whom instead both alleles of PIG-T (another gene involved in glucosylphosphatidylinositol [GPI] biosynthesis) have inactivating mutations, one in the germ line and one somatic.1
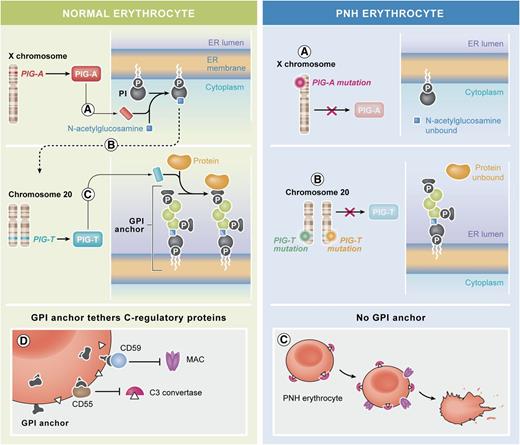
PNH arises through a block in the biosynthesis of the GPI anchor. (Left) Normal erythrocytes and the biosynthesis of GPI. (A) With phosphatidylinositol (PI) on the cytoplasmic side of the ER, the first biosynthetic step, the addition of N-acetylglucosamine to PI is catalyzed by an enzyme complex that comprises a subunit encoded by the X-linked gene PIG-A. (B-C) The biosynthetic pathway proceeds through several enzymatic steps (broken arrow) that add further sugar moieties (green symbols), until the complete GPI molecule is on the luminal side of the ER. Preformed protein (ochre) becomes covalently linked to the GPI anchor through a transamidation reaction. This step is catalyzed by an enzyme complex that comprises a subunit encoded by the gene PIG-T on chromosome 20. (D) Among the red cell proteins that are tethered to the membrane through GPI anchors, ≥2 are important in protecting red cells from activated complement: CD55 that regulates the C3 convertase and CD59 that impedes the lytic action of the membrane attack complex (MAC). (Right) Abnormal PNH erythrocytes: the tethering to the membrane of proteins that need the GPI anchor can be compromised by failure of any of the enzyme reactions above. (A) In the majority of patients with PNH, a somatically acquired inactivating mutation11 of PIG-A (of which in each cell there is only 1 active allele on the X-chromosome) blocks the very take-off of the GPI biosynthesis. (B) Krawitz et al report for the first time a patient in whom the block is instead at the very last step: the GPI anchor is ready, but the protein does not become linked to it because of mutations in both alleles of the PIG-T gene. (C) PNH erythrocytes, lacking CD55, bind C3 convertase; moreover, because they lack CD59, they are vulnerable to MAC attack, and when this takes place, they are lysed. The elements of this figure pertaining to the GPI biosynthetic pathway (top two and middle two panels) were redrawn from the work of Kinoshita et al.,12 and the elements pertaining to the action of complement on normal and on PNH red cells (bottom two panels) were redrawn from Luzzatto et al.13 Professional illustration by Xavier Studio.
PNH arises through a block in the biosynthesis of the GPI anchor. (Left) Normal erythrocytes and the biosynthesis of GPI. (A) With phosphatidylinositol (PI) on the cytoplasmic side of the ER, the first biosynthetic step, the addition of N-acetylglucosamine to PI is catalyzed by an enzyme complex that comprises a subunit encoded by the X-linked gene PIG-A. (B-C) The biosynthetic pathway proceeds through several enzymatic steps (broken arrow) that add further sugar moieties (green symbols), until the complete GPI molecule is on the luminal side of the ER. Preformed protein (ochre) becomes covalently linked to the GPI anchor through a transamidation reaction. This step is catalyzed by an enzyme complex that comprises a subunit encoded by the gene PIG-T on chromosome 20. (D) Among the red cell proteins that are tethered to the membrane through GPI anchors, ≥2 are important in protecting red cells from activated complement: CD55 that regulates the C3 convertase and CD59 that impedes the lytic action of the membrane attack complex (MAC). (Right) Abnormal PNH erythrocytes: the tethering to the membrane of proteins that need the GPI anchor can be compromised by failure of any of the enzyme reactions above. (A) In the majority of patients with PNH, a somatically acquired inactivating mutation11 of PIG-A (of which in each cell there is only 1 active allele on the X-chromosome) blocks the very take-off of the GPI biosynthesis. (B) Krawitz et al report for the first time a patient in whom the block is instead at the very last step: the GPI anchor is ready, but the protein does not become linked to it because of mutations in both alleles of the PIG-T gene. (C) PNH erythrocytes, lacking CD55, bind C3 convertase; moreover, because they lack CD59, they are vulnerable to MAC attack, and when this takes place, they are lysed. The elements of this figure pertaining to the GPI biosynthetic pathway (top two and middle two panels) were redrawn from the work of Kinoshita et al.,12 and the elements pertaining to the action of complement on normal and on PNH red cells (bottom two panels) were redrawn from Luzzatto et al.13 Professional illustration by Xavier Studio.
Paroxysmal nocturnal hemoglobinuria (PNH) has been recognized as a severe hemolytic anemia since the classic paper by Paul Strübing in the 1880s, and it was the first example of a nonneoplastic clonal disorder of hematopoiesis.2 In the 1980s, it became clear that the underlying biochemical defect must be in the biosynthesis of glucosylphosphatidylinositol (GPI), the glycolipid that anchors ∼150 proteins to the cell membrane. Because the biosynthetic pathway is complex (see figure), comprising no less than 10 enzymatic steps, one might have expected to find a defect in any of the genes encoding the respective enzymes. However, somatic cell hybridization studies suggested that the gene defective in PNH was X-linked,3 and by an elegant expression cloning approach, Kinoshita’s group revealed that the defective gene was PIG-A,4 the only gene among those involved in the GPI biosynthesis that maps to the X chromosome (see figure). Thus, 1 somatic hit would be sufficient to cause its inactivation: in fact, a PIG-A mutation was identified in most patients, and it was assumed that in the others perhaps the mutation had been missed.
Now, Krawitz et al report an exceptional patient with PNH who has an intact PIG-A gene. In contrast, both alleles of the PIG-T gene on chromosome 20 have inactivating (null) mutations: one in the germ line is a splicing mutation that entails loss of 28 amino acids, and the other is a somatic deletion of the entire gene. This stimulating exception confirms the rule: the risk of PNH depends on the failure of GPI biosynthesis, and this is true regardless of where along the biosynthetic pathway the block is located. Interestingly, another recent paper5 reported on 4 children (from 2 consanguineous marriages within the same family) who are homozygous for a missense mutation (T183P) in PIG-T. These children have multiple malformations and intellectual disability, but they do not seem to have PNH, presumably because their PIG-T mutation is hypomorphic rather than a null mutation.
The patient reported by Krawitz et al brings up some intriguing issues. First, she had a long history of severe urticarial disease of unknown origin before developing PNH, and once she had established PNH, she also developed severe bowel inflammatory disease with a stormy clinical course (see supplemental Data in Krawitz et al): both of these conditions are thought to be related to immune disturbances, and several immunological abnormalities have been observed in mice with a GPI(−) lymphoid system.6 Second, we do not know whether, before she acquired her somatic PIG-T mutation, the expression of GPI-linked proteins in this patient was normal or whether there was haploinsufficiency: this could be established by testing relatives, which has not been done yet (perhaps for reasons of privacy), and therefore we do not have formal genetic proof that the mutation is inherited. Third, whereas PIG-A is involved in the very first step of GPI biosynthesis, PIG-T is involved in the very last step in the pathway; thus, although the cells have a GPI(−) phenotype, they would have in the endoplasmic reticulum (ER) a complete GPI molecule (see figure). At first sight, this may not favor the notion7 that expansion of the PNH clone is related to the presence of CD1d-restricted GPI-reactive T cells,8 because one might surmise that PIG-T mutant cells, by virtue of having a complete GPI anchor, would not be spared. However, in this particular case, it is possible that immune selection was not the main mechanism of expansion of the PNH clone; alternatively, perhaps the form of GPI against which the T cells are most reactive is displayed within CD1d only after the GPI has been linked to protein.
With respect to genetic lesions of GPI biosynthesis, rare inherited hypomorphic mutations of PIG-A have been found to cause not PNH, but severe congenital abnormalities and early death.9 Inherited hypomorphic mutations of PIG-M have been associated with seizures and thrombosis,10 but not with hemolytic anemia. Inherited hypomorphic mutations of other genes involved in GPI biosynthesis have also given various congenital abnormalities but not PNH (see references in Krawitz et al). Thus, the patient reported by Krawitz et al represents, unfortunately for her, probably the only way that PNH can be (partially) inherited: indeed, she had a severe inactivating mutation of PIG-T in her germ line. However, this patient does not break the paradigm whereby PNH is an acquired disease, as she then suffered a different but equally severe somatic mutation event of her remaining PIG-T allele. It will be important to investigate the clinical state of those of her relatives, if any, who do not have a PIG-T somatic mutation but who share her germ-line PIG-T mutation. This brings up the possibility that mutations of any of the GPI biosynthetic genes, provided they are not frankly haploinsufficient, might be lurking in the heterozygous state in normal people or perhaps they might give phenotypes different from PNH. One also wonders how often, on systematic sequencing, one may find PNH associated with non–PIG-A mutations.
Conflict-of-interest disclosure: The author declares no competing financial interests.