In this issue of Blood, Li et al reveal the genetic elements that control the activity of Bcl11b, a critical regulator of T-cell development. Lineage-defining transcription factors (TFs), such as Bcl11b, control key steps in cellular differentiation throughout development, and understanding how these TFs are themselves regulated represents a major challenge.1
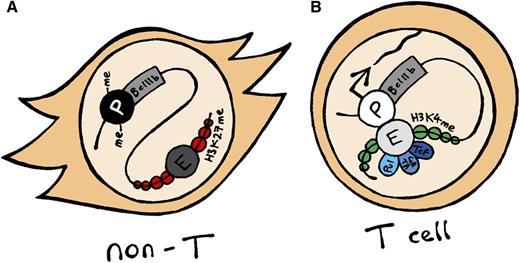
Cartoon depicting the Bcl11b locus in non-T and T cells. (A) In non-T cells, the Bcl11b promoter is often DNA-methylated and does not physically interact with its downstream enhancer, which is marked by H3K27me3. (B) In T cells and committed T-cell precursors, the actively transcribing Bcl11b promoter is unmethylated and interacts with the enhancer, which is marked by H3K4me2 (and H3K4me1) and bound by transcription factors associated with T-cell development such as Pu1, Gata3, and Tcf1 (among others).
Cartoon depicting the Bcl11b locus in non-T and T cells. (A) In non-T cells, the Bcl11b promoter is often DNA-methylated and does not physically interact with its downstream enhancer, which is marked by H3K27me3. (B) In T cells and committed T-cell precursors, the actively transcribing Bcl11b promoter is unmethylated and interacts with the enhancer, which is marked by H3K4me2 (and H3K4me1) and bound by transcription factors associated with T-cell development such as Pu1, Gata3, and Tcf1 (among others).
Expression of many developmentally important genes is associated with the activities of distal enhancers, which often each drive target gene expression in distinct, specific cell types.2 Particularly in vertebrates, these enhancers can be separated by large genomic intervals from their target promoter(s), and their characterization is nontrivial: in consequence, only a few studies have succeeded in identifying long-range enhancers that are both necessary and sufficient for gene expression in defined cell types.3
The development of T cells from hematopoietic precursors is a well-studied model system for cell differentiation. In this process, Bcl11b plays a pivotal role as a commitment factor: it is required for repression of alternate lineages in developing progenitors, after initial T-cell specification by other TFs.4 Bcl11b orthologs are found in all vertebrate genomes, suggesting that Bcl11b may have evolved together with lymphocyte lineages. Genomic translocations downstream of Bcl11b are common in some T-cell tumors, suggesting the existence of cis-regulatory sequences.5 Nevertheless, the mode of Bcl11b regulation in hematopoietic cells has until now not been determined.
In the present study, Li et al initially use cell-type–specific DNA methylation, histone modifications, and TF binding as clues to locate candidate regulatory elements.1 The first is the Bcl11b promoter itself: this contains an overlapping CpG island which is unmethylated in T cells, yet methylated in myeloid-lineage cells, mirroring its cell-type–specific activity. The authors then pinpoint a putative enhancer (termed “Major Peak”), located 850 kb downstream of the Bcl11b promoter in an adjacent gene desert, which is bound by multiple TFs involved in T-cell development and which physically interacts with the Bcl11b promoter specifically in T cells (see figure). This enhancer additionally interacts with a region within Bcl11b exon 3 also bound by Ikaros and Tcf1, but, critically, not with other nearby (Vrk1) or T-cell–specific (Cd3e) genes.
Li et al next use functional reporter assays to demonstrate that the Bcl11b promoter and enhancer are together sufficient to direct gene expression in T cells, but not in non–T-lineage hematopoietic cells. Distinct subregions within the enhancer function additively: a pair of Tcf1-bound sites, plus a separate highly conserved region occupied by multiple TFs, are individually sufficient to form a weak, broadly functional enhancer, and in combination can impart T-cell specificity; but the presence of an additional terminal region (found only in mammals and lacking obvious TF binding) is required for full activity. Finally, the authors elegantly use a modified BAC transgene to show that this enhancer is also necessary for expression of Bcl11b from its local genomic locus in T cells.
Altogether, these experiments define the Major Peak enhancer in conjunction with the Bcl11b promoter region as key regulatory elements controlling Bcl11b activation in T cells. Of course, there may be additional undiscovered elements within the 1.8-Mb gene desert adjacent to Bcl11b which contribute to its expression pattern, and only deletion of the enhancer in animals could definitively address this. Likewise, the fine specificities of the Bcl11b promoter and enhancer remain to be functionally tested at individual T-cell developmental stages, or outside of the hematopoietic system.
An intriguing feature of the Bcl11b enhancer is its promoter specificity: in T cells, it interacts only with the Bcl11b promoter, and in functional assays it cannot drive transcription from a heterologous SV40-derived promoter. This may be a general feature for extremely long-range enhancers, to avoid unwanted activation of nontarget promoters. However, the Bcl11b enhancer encompasses DNase-hypersensitive sites which were previously shown to activate juxtaposed genes after genomic translocation,5 implying that promoter specificity is either incomplete, or lost during leukemic transformation.
At first glance, the finding that the Bcl11b promoter is unmethylated in T cells but not in myeloid cells might suggest that activation could accompany a T-cell–specific loss of DNA methylation. However, genome-wide bisulphite-sequencing data in mice6 and humans indicate that the Bcl11b promoter is unmethylated both in embryonic stem cells and various other nonhematopoietic cell types: thus, an alternative scenario could be that DNA methylation is acquired during hematopoietic differentiation to limit Bcl11b expression in particular lineages.
Armed with the newly revealed identities of its regulatory sequences, the task ahead will be to determine how these activate Bcl11b specifically at the DN2b stage of T-cell development. Many TFs which bind the Bcl11b enhancer (and promoter) are also involved in the specification of lymphocyte and T-cell lineages at earlier developmental stages,4 including Ikaros, Pu1, Tcf1, Gata3, and Runx1. Thus, the same TFs which combine to drive T-cell differentiation also contribute, through regulation of Bcl11b, to repression of alternative cell fates: this recalls the analogous regulation of Pax5 by Ebf1 during B-cell differentiation.7 However, although these TFs may be involved in establishment of a functional enhancer (and this role has been proposed for Pu1 in other cell types),8 they cannot alone explain the precise onset of Bcl11b expression.
One emerging notion is that the chromatin status of enhancers can contribute to their patterns of activity.9,10 Accordingly, the Bcl11b enhancer undergoes loss of H3K27me3 at the DN2b stage, concomitant with the onset of target gene expression; it is also embedded in an extended region bearing high levels of H3K9me3 in many nonhematopoietic cell types. Further work will be required to determine whether either mark causatively influences the Bcl11b enhancer, and how each is established and regulated at this locus. It is notable, though, that Bcl11b is located adjacent to a syntenic 1- to 2-Mb gene desert in all vertebrate species, and its conserved enhancer sequences invariably lie within this. Gene deserts were previously proposed to represent favored sites for regulatory sequences of developmental genes3 ; possibly, this placement may enable regulation by chromatin modifications while minimizing adverse impact on nearby gene expression.
In summary, the study by Li et al nails down the key regulatory elements that dictate Bcl11b expression in T cells, and opens the door for future experiments to dissect how expression of this lineage-defining vertebrate TF can itself be regulated during development.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal