Key Points
Salmonella is engineered to specifically infect tumor cells based on recognition of a tumor antigen by a bacterial-expressed antibody.
Once inside, Salmonella can transfer cytotoxic cargos to destroy human tumors even in the absence of an adaptive immune response.
Abstract
Escape from immune detection favors both tumor survival and progression, and new approaches to circumvent this are essential to combat cancers. Nonvirulent, tumortropic bacteria, such as Salmonella typhimurium, can unmask a tumor by transforming it into a site of inflammation; however, the nonspecific invasiveness of Salmonella leads to off-target effects diluting its therapeutic efficacy and making its use in human patients inherently risky. Here, we demonstrate that Salmonella tumor specificity can be significantly improved via a surface-expressed single-domain antibody directed to a tumor-associated antigen (CD20). Antibody-dependent bacterial targeting specifies the infection of CD20+ lymphoma cells in vitro and in vivo, while significantly diminishing nonspecific cell invasion. Indeed, CD20-targeted Salmonella was less generally invasive, even in organs that normally serve as physiological reservoirs. Furthermore, tumor-specific Salmonella engineered to carry the herpes simplex virus thymidine kinase prodrug-converting enzyme effectively treats human lymphoma xenografts when coadministered intratumorally or intravenously with ganciclovir in mice lacking a functional adaptive immune system. Therefore, tumor-targeted Salmonella could prove effective even in those patients displaying a debilitated immune system, which is often the case with late-stage cancers. Altogether, antibody-displaying Salmonella vectors can mediate a tumor-specific response and rejection with few detectable adverse effects while specifically delivering cytotoxic payloads.
Introduction
Developing tumors are scrutinized by an immune system capable of recognizing and destroying aberrant cells before they become pathologic. To survive, a tumor must adopt a number of well-studied immune evasion mechanisms that overcome this monitoring.1 Cancer immunotherapy seeks to harness the body’s immune response to treat tumors despite immune evasion adaptations. One interesting and innovative approach uses tumor-colonizing bacteria to counteract the immunosuppressive microenvironment. Indeed, a number of bacterial strains, including Clostridium, Bifidobacterium, and Salmonella, exhibit a remarkable tropism for tumors, indicating a therapeutic potential (reviewed by Forbes).2 Although the precise mechanisms facilitating bacterial tumor colonization are unclear, both the leakiness of the tumor vasculature and the hypoxic conditions found therein contribute.
Attenuated strains of Salmonella typhimurium represent valuable models for in vivo use, being genetically well characterized, easy to manipulate, and approved for administration in humans.3 Our group,4,5 among others,6,7 has previously demonstrated that Salmonella can eradicate tumors in mouse models. Indeed, intratumoral injection of Salmonella into aggressive melanomas has led to tumor regression and rejection. Furthermore, bacterial injection into primary tumors has caused significant growth retardation of untreated, distal tumors from Salmonella-dependent secondary immune responses created against tumor-associated antigens.4 This was reliant on the Salmonella-dependent upregulation of connexin43, a transmembrane protein involved in gap junction (GJ) formation.8 GJ between tumor cells and dendritic cells (DCs) favored tumor antigen crosspresentation and the establishment of an anti-tumor immune response.
In this study, we aimed to engineer Salmonella to reduce its systemic effects, while retaining its antitumor activity and potentiating its immune system–independent cytotoxic effects. Anti-tumor antibodies used alone or to convey cytotoxic cargoes (radiolabels, prodrug-converting enzymes) represent frontline therapies for many cancers.9-11 We hypothesized that anti-tumor antibody expression would enhance Salmonella tumor cell invasion while disrupting general cell invasion. As a proof-of-concept experiment, we selected and characterized a camelid single-domain (VHH) antibody against the human CD20 (hCD20) tumor antigen from a phage-display library12 and expressed it on Salmonella. CD20, the antigen targeted in rituximab antibody immunotherapy of non-Hodgkin and diffuse large B-cell lymphomas,10,13 represented an ideal target, as it was a well-studied tumor-associated antigen.
Here, we show success not only in specifically targeting tumor cells but also in delivering within them, via Salmonella infection, the herpes simplex virus thymidine kinase (HSV-TK). Thus, tumor cells became ganciclovir sensitive, thereby potentiating the therapeutic effect of Salmonella even in the absence of adaptive immunity.
Methods
Phage panning and screening
B16-F10 murine melanoma cells were transfected with either hCD20-expressing or empty vector Babe-IRES-Puro (BIP) plasmid. Cells were harvested after 2 days, and hCD20 expression was verified by a fluorescence-activated cell sorter (FACS) using a rabbit anti-hCD20 antibody (Abcam). A total of 5 × 106 B16F10-BIP empty vector cells were incubated with 1 × 1015 transforming units of the COGENTECH 1–naïve camelid VHH bacteriophage display library.12 Cells were pelleted, and supernatant (containing unbound phage) was added to 5 × 106 B16F10-BIP-hCD20 cells. Phage elution, propagation, quantification and purification, and actual phage transforming units were performed as described previously,12 and output phage was used for second and third pannings.
A total of 200 third-round B16F10-hCD20–interacting phage colonies were picked, and VHH-pIII-protein–containing periplasms were produced as described previously.12 Clones were screened in quadruplicate by dot blot vs either CD20+ Namalwa B lymphoma or CD20− 697 B leukemia cells. Binding was detected via anti-HA 12CA5 Ab [1 μg/mL] and goat anti-mouse-HRP Ab, followed by enhanced chemiluminescence development (GE Healthcare Life Sciences), and was quantified using ImageJ software. A total of 17 clones most consistently binding CD20+, and not CD20−, cells were sequenced, revealing repeated VHHs (5/17), of which clone 2G9 (3/17) alone could be purified.
VHH purification/characterization
2G9- and glutathione S-transferase (GST)b1-VHH were cloned into pHEN6 and VHH purified as described previously.12 FACS verifications of VHH-binding specificities were detected via anti-His mAb (Santa Cruz) followed by sheep anti-mouse-biotin IgG-PE antibodies (Sigma-Aldrich) and were compared with cells probed with rabbit anti-CD20 antibody (Abcam), followed by rat anti-rabbit AlexaFluor546 (Molecular Probes).
2G9-CD20 protein interaction was verified by coincubating VHH with Namalwa cell membrane fractions (Native Membrane Protein Isolation kit [Calbiochem]). Antibodies were pulled down (nickel nitrilotriacetic acid agarose [VHH] or Protein A-Sepharose [rAb]), washed, and boiled into Laemmli buffer for western blot analysis. The presence of CD20 in the immunoprecipitates was visualized as described above.
Bacteria and plasmids
All experiments used the SL3261 aroA–deficient S typhimurium (AT-Salmonella)14 as described previously.4,5 Colony-forming units (cfus) were determined for each transformant with or without isopropyl-L-thio-beta-D-galactopyranoside induction and were verified after each experiment.
hCD20 was amplified by polymerase chain reaction from Ramos human B lymphoma cDNA, sequence verified, and cloned into the BIP (retroviral)15 and 945-green fluorescent protein (GFP) (lentiviral) vectors (Dr L. Naldini, Telethon Institute of Gene Therapy, San Raffaele Scientific Institute, Milan, Italy).16 The HSV-TK gene from pHSV-TK (Dr V. Russo, Department of Biological and Technological Research, San Raffaele Scientific Institute)17 was cloned into pET15b plasmid. VHHs were cloned into the pHEN612 and pVUB418 vectors. Primer sequences and detailed cloning strategies are available on request.
Outer membrane translocation of VHH-ompA fusion proteins were routinely verified by dot blot of intact bacteria and were detected by western blotting. Bacterial outer membrane protein fractionation was performed as described previously,19 and VHH was similarly detected.
Cells
B16F10 cells were cultured as described previously,4 as were the MCA203 fibrosarcoma20 and the CT26 colon carcinoma21 cells (Dr Mario Colombo, Immunotherapy and Gene Therapy Unit, Istituto Nazionale dei Tumori, Milan, Italy).
hCD20-945-GFP and 945-GFP empty vector lentivirus-containing supernatants were produced, and infections were performed following standard protocols. FACS verification of hCD20-expression was performed as described above.
Immunofluorescence
Cells were counted, seeded onto Permanox chamber slides (Laboratory-Tek), and coincubated with Salmonella at defined multiplicities of infection (MOIs) for 2 hours at 37°C. Slides were then subjected to immunofluorescence staining to discriminate extracellular (doubly positive for Cy5- and Cy3-secondary antibodies) and intracellular bacteria (Cy5-singly-positive)22 using an anti-Salmonella antibody (Virostat). Cytoplasm was visualized via phalloidin-fluorescein isothiocyanate (FITC) and nuclei by 4′,6 diamidino-2-phenylindole. Images were taken on an Olympus AX70 fluorescent microscope. Quantification of extracellular and intracellular bacteria was performed in a blinded manner.
In vivo murine tumor treatments
Six-week-old female C57/Bl6J-OlaHsd mice (Harlan Laboratories) were injected subcutaneously with 5 × 105 MCA203-empty vector or -CD20-GFP cells in contralateral flanks. Tumors were grown until the desired dimensions were reached: short-term analyses 5 to 8 mm2; long-term analyses 3 to 4 mm2.
For biodistribution experiments, tumor-bearing mice received a single intravenous injection of 5 × 106 cfus of the indicated Salmonella. Animals were euthanized 24 hours later; liver, spleen, tumor, bone marrow, and brain were excised, minced, and digested with collagenase D (Roche). Total bacterial load per organ was quantified by serially diluting counted cells. Intracellular bacteria were determined in parallel by a gentamicin-protection assay (50 μg/mL, 1 hour, 37°C).
For short-term treatments, mice were either immunized or were not immunized vs Salmonella via oral gavage19 and then received intravenous 5 × 106 cfus of the indicated Salmonella. Tumor dimensions were measured every 2 days for up to 2 weeks.
For long-term survival analyses, 1 × 108 cfus were administered intratumorally into nonimmunized mice. Tumor dimensions were measured as above, and mice were monitored for up to 6 months. Mice were judged tumor-free when no mass was palpable for 1 month minimum. Surviving mice were rechallenged with 5 × 105 MCA203 cells at a distal site, and tumor development was monitored for 3 months.
In vivo treatment of human xenografts
Five-week-old, female nonobese diabetic/severe combined immunodeficiency (NOD/SCID) mice (Charles River) were injected subcutaneously with either 5 × 106 Namalwa or Karpas299 cells.23 Bacterial biodistribution was determined as detailed above. When tumors had grown sufficiently, mice received 1 × 108 cfus of bacteria intratumorally once per week for 2 weeks, and tumor growth was tracked as mentioned.
For ganciclovir sensitization experiments, Salmonella containing both the pVUB4KAN (with or without VHH) and pET15b-HSV-TK plasmids was selected. HSV-TK expression was verified using goat anti-HSV-TK antibody (SantaCruz). Bacteria were injected either intratumorally (1 × 108 cfus) or intravenously (5 × 106 cfus) into tumor-bearing animals every 4 days until the end of the experiment; control animals received phosphate-buffered saline (PBS) alone. Concomitantly, mice were injected intraperitoneally with 50 mg/kg of ganciclovir every 12 hours for the duration of the experiment (100 mg/kg/day).24 Tumor growth was monitored every 2 days. Survivors partially or fully rejecting tumors were treated for 1 additional month.
All animal studies were approved by the Institutional and Italian Health Ministries Animal Care and Use Committees. All mice were maintained and handled in accordance with local and international laws and relevant regulations.
Results
Engineering Salmonella to express a recombinant antibody against the hCD20 tumor-associated antigen
The tropism of Salmonella for tumors has been well documented.3,25,26 Although usage of attenuated strains, such as the AT14 mutant, significantly reduces acute effects,27 its widespread, general cell invasion dilutes its therapeutic efficacy.26,28 We hypothesized that Salmonella could be engineered to become tumor specific via the bacterial surface overexpression of an antibody against a tumor-associated antigen. Thus, antigen-expressing cells (tumors) would be invaded, but the antigen-negative cells (healthy tissue) would not. As a proof-of-principle experiment, we chose the hCD20 protein, the target of the rituximab anti-lymphoma antibody immunotherapy.10,13 To specifically fish out CD20-directed VHH and not general lymphoma-associated targets, we transformed B16F10 melanoma cells to ectopically express hCD20. Specific VHHs recognizing hCD20 in its native conformation were isolated by panning a preimmune phage-display library12 against intact cells. Despite poor protein expression by the B16F10 cells (7% hCD20-positivity [supplemental Figure 1A]), we observed a significant enrichment of recovered phages after second and third rounds of panning (13-fold and 1.9-fold, respectively; data not shown), compared with negative control pannings. A total of 200 clones were then selected and were screened, as described in the “Methods” section. The hCD20-specificity of a selected number of clones (n = 17) was confirmed via FACS, examples of which can be seen in Figure 1A and supplemental Figure 1B. Sequencing revealed 2 VHH-sequences present in multiple clones, and they were subcloned and purified for further characterization. Because of its biochemical stability, we selected the VHH encoded by clones 2G9/1H5/2B10 (2G9), which was able to distinguish by FACS, compared with a GST-binding control VHH (GSTb1), CD20+ lymphomas from CD20- leukemias (Figure 1B), or CD20− T lymphomas (Karpas299; data not shown), and effectively immunoprecipitated hCD20 protein from cell lysates (Figure 1C).
Characterization of an anti-CD20 camelid VHH and its expression on the Salmonella surface. (A) Fluorescence activated cell sorting dot plots of Namalwa cells stained with periplasmic lysates obtained from the indicated VHH-phage-producing E coli clones compared with a secondary/tertiary antibody negative control (Ab control) and an anti-CD20 monoclonal antibody positive control (CD20 mAb control). (B) Quantification of cell binding as determined via FACS using highly purified 2G9-6xHis-VHH or GSTb1-6xHis-VHH incubated with Namalwa CD20+ or 697 CD20− cells. (C) Immunoprecipitation of native CD20 from intact cell membranes with either anti-CD20 rabbit polyclonal antibody (positive control), GSTb1-VHH negative control, or 2G9-VHH, as compared resin-only controls (Protein-A Sepharose or nickel nitrilotriacetic acid) with a lane containing the total protein before precipitation. (D) VHH expression on AT-Salmonella outer membrane fractions detected via the 6xHis tag (2G9 or GSTb1) or not (ompA empty vector) derived from equal cfus of bacterial pellets.
Characterization of an anti-CD20 camelid VHH and its expression on the Salmonella surface. (A) Fluorescence activated cell sorting dot plots of Namalwa cells stained with periplasmic lysates obtained from the indicated VHH-phage-producing E coli clones compared with a secondary/tertiary antibody negative control (Ab control) and an anti-CD20 monoclonal antibody positive control (CD20 mAb control). (B) Quantification of cell binding as determined via FACS using highly purified 2G9-6xHis-VHH or GSTb1-6xHis-VHH incubated with Namalwa CD20+ or 697 CD20− cells. (C) Immunoprecipitation of native CD20 from intact cell membranes with either anti-CD20 rabbit polyclonal antibody (positive control), GSTb1-VHH negative control, or 2G9-VHH, as compared resin-only controls (Protein-A Sepharose or nickel nitrilotriacetic acid) with a lane containing the total protein before precipitation. (D) VHH expression on AT-Salmonella outer membrane fractions detected via the 6xHis tag (2G9 or GSTb1) or not (ompA empty vector) derived from equal cfus of bacterial pellets.
To create Salmonella displaying 2G9 on its outer membranes, the VHH was cloned into the pVUB4 vector, fusing it to the ompA transmembrane protein domain.18 The isopropyl-L-thio-beta-D-galactopyranoside-induced expression of 2G9 on the outer surface of AT-Salmonella was visualized using purified membrane fractions (Figure 1D), and VHH translocation was thereafter monitored by dot blot (data not shown). As a final confirmation, 2G9-Salmonella efficiently bound hCD20+ Namalwa cells, although failing to bind CD20− 697 cells, as visualized by immunofluorescence (supplemental Figure 2).
2G9-Salmonella exhibits enhanced binding and entry into CD20-expressing cells in vitro
Next, we determined whether VHH expression could direct bacterial invasion to cells expressing the targeted marker on their membranes, as AT-Salmonella remains competent to invade murine cells. We generated mouse colon carcinoma (CT-26) cells (Figure 2A), and fibrosarcoma (MCA203) cells (images not shown) to stably express hCD20. CD20+ and CD20- WT counterpart cells were coincubated with AT-Salmonella expressing no VHH, 2G9-VHH, or the GSTb1-VHH at defined MOIs. Salmonella localization was then examined via immunofluorescence under conditions allowing for the discrimination of intracellular and extracellular bacteria, based on differentially staining before and after permeabilization.22 As shown in Figure 2A-B, 2G9-Salmonella bound and invaded the CD20+ cells at least as efficiently as unmodified AT-Salmonella, but significantly more than GSTb1-Salmonella (displaying the irrelevant control VHH). 2G9-Salmonella bound cells 5.6-fold (MCA203) (P = .006) and 7.9-fold better (CT26) (P = 3 × 10−9) than the GSTb1 control (Figure 2B). As hypothesized, that the surface-expression of VHH on Salmonella may restrict its infectivity, expression of GSTb1-VHH inhibited the capacity of Salmonella to bind and invade tumor cells, independent of the presence of CD20. Indeed, bound 2G9-Salmonella were significantly more invasive than GSTb1-Salmonella (64% vs 29% for MCA203 cells [P = .0007] and 49% vs 16% for CT26 cells [P = 7 × 10−6]) (Figure 2C). The cfus for each strain were verified postinfection (Figure 2D), thus eliminating the possibility that the observations were the result of differing amounts of bacteria present. Altogether, these results suggest that bacterial surface expression of recombinant antibodies could disrupt nonspecific cell interactions of Salmonella while the antibody-mediated recognition of a cellular marker preserves cellular binding and invasion.
2G9-Salmonella efficiently interacts with and invades CD20-expressing cells. (A) Representative immunofluorescence images of CD20-expressing CT26 cells infected with control AT-Salmonella (left panel), 2G9-Salmonella (middle panel), or GSTb1-Salmonella (right panel) at MOI 10, showing extracellular bacteria (Cy5-positive [blue]), intracellular bacteria (Cy3-Cy5-double positive [red]), and cytoplasm/phalloidin (FITC [green]) staining. Images shown are representative of 3 separate immunofluorescence infection experiments comprising at least 5 randomly selected fields of view per infection. (B) Quantification of total number of bound per infection (extracellular + intracellular), determined from at least 4 immunofluorescence images per infection, normalized per the cell count in field of view. (C) Quantification of invasiveness of bound bacteria (intracellular/[intracellular + extracellular]). (D) Total number of bacterial cfus used in (A) and (B), as determined by limiting dilution assay of the bacteria used for infection. All images shown were taken with a 40× objective.
2G9-Salmonella efficiently interacts with and invades CD20-expressing cells. (A) Representative immunofluorescence images of CD20-expressing CT26 cells infected with control AT-Salmonella (left panel), 2G9-Salmonella (middle panel), or GSTb1-Salmonella (right panel) at MOI 10, showing extracellular bacteria (Cy5-positive [blue]), intracellular bacteria (Cy3-Cy5-double positive [red]), and cytoplasm/phalloidin (FITC [green]) staining. Images shown are representative of 3 separate immunofluorescence infection experiments comprising at least 5 randomly selected fields of view per infection. (B) Quantification of total number of bound per infection (extracellular + intracellular), determined from at least 4 immunofluorescence images per infection, normalized per the cell count in field of view. (C) Quantification of invasiveness of bound bacteria (intracellular/[intracellular + extracellular]). (D) Total number of bacterial cfus used in (A) and (B), as determined by limiting dilution assay of the bacteria used for infection. All images shown were taken with a 40× objective.
VHH overexpression diminishes generic Salmonella invasion in vitro
We then confirmed that CD20-targeted Salmonella lost the capacity to bind or invade cells lacking CD20, via infections performed with CD20− MCA203 (Figure 3A) and CT-26 (Figure 3B) cells. Although wild-type AT-Salmonella entered both CD20+ and CD20− cells with similar efficiency (compare Figure 2A with Figure 3A-B, left panels), 2G9-Salmonella predominantly invaded the CD20+ cells (Figure 2A-B and Figure 3A-B). Indeed, when encountering cells lacking their targeted antigen, 2G9-Salmonella mimics GSTb1-Salmonella, becoming incapable of binding or invading cells (Figure 2A-B and Figure 3A-B). Consequently, the AT-Salmonella bound the MCA203 and CT-26 cells 4.4-fold (P = .01) and 1.8-fold (P = .01) better, respectively, than its 2G9-expressing counterpart (Figure 3C). AT-Salmonella also invaded better (67% vs 33% [MCA203, P = .02] and 59% vs 21% [CT26, P = .002], AT vs AT-2G9, respectively) (Figure 3D) when using comparable cfus in each infection (data not shown). These data confirm our hypothesis that overexpression of an antibody at the Salmonella surface confines binding and invasion only to cells expressing the targeted antigen.
VHH-expressing Salmonella do not significantly invade CD20− cells. Representative immunofluorescence images of empty vector–transfected (A) MCA203 and (B) CT26 cells infected with control AT-Salmonella (left panels) or 2G9-Salmonella (right panels) at MOI 10, showing extracellular bacteria (Cy5-positive [blue]), intracellular bacteria (Cy3-Cy5-double positive [red]), and cytoplasm/phalloidin (FITC [green]) staining. Images shown are representatives of 3 separate immunofluorescence-infection experiments comprising at least 4 randomly selected fields of view per infection. (C) Quantification of total number of bound per infection (extracellular + intracellular), determined from 4 immunofluorescence images per infection, normalized per the cell count in field of view. (D) Quantification of invasiveness of bound bacteria (intracellular/[intracellular + extracellular]) shown in Figure 3C.
VHH-expressing Salmonella do not significantly invade CD20− cells. Representative immunofluorescence images of empty vector–transfected (A) MCA203 and (B) CT26 cells infected with control AT-Salmonella (left panels) or 2G9-Salmonella (right panels) at MOI 10, showing extracellular bacteria (Cy5-positive [blue]), intracellular bacteria (Cy3-Cy5-double positive [red]), and cytoplasm/phalloidin (FITC [green]) staining. Images shown are representatives of 3 separate immunofluorescence-infection experiments comprising at least 4 randomly selected fields of view per infection. (C) Quantification of total number of bound per infection (extracellular + intracellular), determined from 4 immunofluorescence images per infection, normalized per the cell count in field of view. (D) Quantification of invasiveness of bound bacteria (intracellular/[intracellular + extracellular]) shown in Figure 3C.
2G9-Salmonella preferentially invades and rejects CD20+ tumors in vivo
Next, we examined the capacity of the 2G9-Salmonella to invade tumors in vivo. Mice were injected subcutaneously in opposite flanks with MCA203 cells infected with either empty vector or hCD20-encoding vector, with the empty vector tumors acting as hCD20-free negative control tissues to confirm antigen-specific targeting or invasion in vivo. When the tumors had grown sufficiently, the mice were injected intravenously with 5 × 106 cfus of either the 2G9-Salmonella or the GSTb1-Salmonella. At 24 hours later, the animals were euthanized and bacterial distribution and invasiveness were quantified via a gentamicin protection assay. Although the total bacterial load within either the CD20+ or CD20− tumor was equivalent, regardless of which VHH was displayed (data not shown), the 2G9-Salmonella accumulated intracellularly 5.2-fold more within the CD20+ tumor than the CD20− tumor within the same animals (Figure 4A) (P = .04), indicating that VHH-targeted Salmonella did not lose tumor tropism and retained antigen-specific invasion in vivo. Importantly, only minimal in vivo invasion of either tumor was detected using GSTb1-Salmonella (Figure 4A) (P < .05). Importantly, compared with mice receiving unmodified AT-Salmonella, VHH-targeted bacteria did not exhibit significant off-target invasion of known in vivo reservoirs (spleen and liver)26 (Figure 4B), did not aberrantly localize elsewhere (data not shown), and did not affect the number of circulating B cells (supplemental Figure 3), thereby confirming the disruption of general, nonspecified cell invasion in vivo.
2G9-Salmonella specifically invades and rejects CD20-positive tumors in vivo, increasing mouse survival duration. (A) Quantification of in vivo intracellular 2G9-Salmonella vs GSTb1-Salmonella within CD20+ and CD20− MCA203 tumors, 24 hours after intravenous delivery (n = 4 per group). (B) In vivo, intracellular organ biodistribution (liver, spleen, and tumor) of unmodified AT-Salmonella vs 2G9-Salmonella 24 hours after intratumoral delivery as assessed by gentamicin protection assay. n.d.= not detectable. (C-D) Tracking of acute-phase tumor growth after a single intratumoral injection of the indicated bacteria into mice bearing both CD20+ and CD20− MCA203 tumors in opposite flanks (n = 5 mice per group [PBS and empty vector-AT-Salmonella] or 8 mice per group [2G9-AT]). (E) Kaplan-Meier curves of mice bearing either CD20+ or CD20− MCA203 tumors injected twice with 2G9-Salmonella or empty vector-Salmonella (n = 6 mice per group). (F) Actual cfus injected as determined after injection by limiting dilution assay of the injected bacterial solution, bars 1 and 2 corresponding to the data presented in (A) and bars 3 and 4 corresponding to the data shown in (C) and (D). All data shown are from representative experiments, which were repeated 3 times, yielding similar results.
2G9-Salmonella specifically invades and rejects CD20-positive tumors in vivo, increasing mouse survival duration. (A) Quantification of in vivo intracellular 2G9-Salmonella vs GSTb1-Salmonella within CD20+ and CD20− MCA203 tumors, 24 hours after intravenous delivery (n = 4 per group). (B) In vivo, intracellular organ biodistribution (liver, spleen, and tumor) of unmodified AT-Salmonella vs 2G9-Salmonella 24 hours after intratumoral delivery as assessed by gentamicin protection assay. n.d.= not detectable. (C-D) Tracking of acute-phase tumor growth after a single intratumoral injection of the indicated bacteria into mice bearing both CD20+ and CD20− MCA203 tumors in opposite flanks (n = 5 mice per group [PBS and empty vector-AT-Salmonella] or 8 mice per group [2G9-AT]). (E) Kaplan-Meier curves of mice bearing either CD20+ or CD20− MCA203 tumors injected twice with 2G9-Salmonella or empty vector-Salmonella (n = 6 mice per group). (F) Actual cfus injected as determined after injection by limiting dilution assay of the injected bacterial solution, bars 1 and 2 corresponding to the data presented in (A) and bars 3 and 4 corresponding to the data shown in (C) and (D). All data shown are from representative experiments, which were repeated 3 times, yielding similar results.
To determine whether antibody-dependent tumor recognition and invasion by Salmonella could be therapeutically beneficial, we orally immunized mice against Salmonella,19 and CD20+ and CD20− MCA203 tumors were intratumorally injected with 1 × 108 cfus of either 2G9- Salmonella or empty vector-Salmonella. 2G9-Salmonella elicited a 50% reduction of CD20+ tumors with respect to their preinjected state, but the CD20− tumors continued to grow (Figure 4C-D, P = .01 at day 10). In contrast, treatment with the empty vector-Salmonella failed to induce tumor regression, which, instead, doubled in size within 10 days of initial treatment of the tumors (Figure 4D), much like the 2G9-injected CD20− tumors. As expected,4 intratumoral Salmonella was itself partially protective, as PBS-injected tumors grew faster than Salmonella-injected tumors (Figure 4D). Empty vector-Salmonella and not GSTb1-Salmonella served as a control because the latter, being incapable of infecting tumors, would have masked the nonspecific anticancer effect of invasive Salmonella, thus confounding the comparison.
Although intratumoral delivery of Salmonella into preimmunized animals was protective in an acute response, whether the 2G9-Salmonella–induced regression of CD20+ tumors could increase survival duration remained uncertain. Nonimmunized mice bearing either CD20+ or CD20− MCA203 tumors were injected intratumorally, twice, 1 week apart, with either the 2G9-Salmonella or empty vector-Salmonella (1 × 108 cfus), and tumor dimensions and mouse survival duration were then tracked. As seen in Figure 4E, overall survival time was significantly extended in only those mice bearing hCD20 tumors and receiving 2G9-Salmonella (log rank P = .02). Indeed, we consistently observed that up to 67% of the CD20+ tumor-bearing mice treated with 2G9-Salmonella survived tumor-free for more than 3 months after the end of the experiment. This rate was significantly better than the rate seen for CD20+ tumors treated with empty vector-Salmonella (12.5%), whereas no mice bearing CD20− tumors exhibited prolonged survival time with either bacterium. Successful tumor rejection generated anti-tumor immunity, as subsequent rechallenge of the CD20+ tumor survivors (n = 6) with MCA203 cells failed to yield secondary tumors (data not shown). The number of cfus used within each condition was verified to be identical (Figure 4F and data not shown). These observations demonstrate that the expression of tumor-antigen–specific VHH on Salmonella improves the selectivity for antigen-bearing tumor cells, culminating in tumor regression, rejection, and the establishment of tumor-protective immunity while simultaneously disrupting nonspecific invasion of healthy tissues.
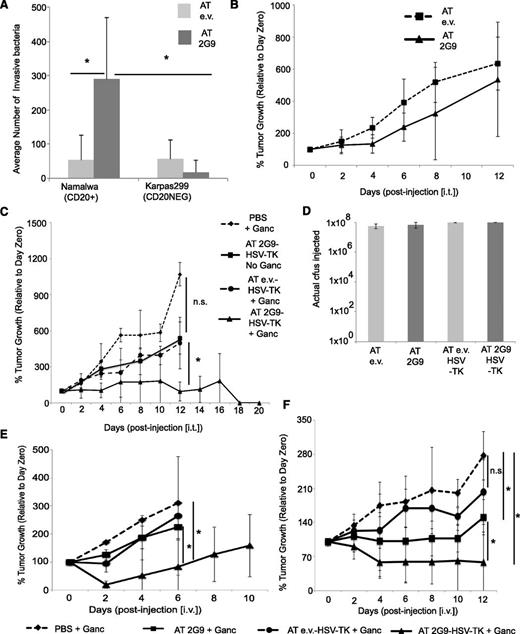
2G9-Salmonella carrying a prodrug-converting enzyme can preferentially invade and destroy hCD20+ lymphoma xenografts
We further sought to extend the applicability of our system by examining whether our tumor-targeting Salmonella could be used to treat human tumors. Therefore, we used the NOD/SCID xenograft model in which the absence of an adaptive immune system allows for human tumor cell outgrowth. CD20+ Namalwa cells or the CD20− Karpas299 were injected subcutaneously in opposite flanks23 and then were injected 3 weeks later intratumorally with 1 × 108 cfus of either 2G9-Salmonella or empty vector-Salmonella. Notably, the 2G9-bacteria invaded the CD20+ tumor cells by an average of 16.9-fold better than the CD20− tumors (P = .03), and 5.5-fold better than the empty vector-Salmonella (P = .05) (Figure 5A). Despite this finding, no significant tumor regression was observed after administration of either the 2G9-Salmonella or empty vector-Salmonella according to the previously outlined immunotherapeutic protocol (Figure 5B). This trend was to be expected, as we had observed previously that the formation of tumor-specific CD8+ T cells, lacking in NOD/SCID animals, was essential for Salmonella-based cancer immunotherapy.4
Human xenograft treatment via 2G9-Salmonella bearing the HSV-TK enzyme. (A) Intracellular 2G9-Salmonella and empty vector-Salmonella measured 24 hours after intratumoral injection into mice bearing both CD20+ and CD20− Karpas299 tumors. (B) Tracking of acute-phase tumor growth after a single intratumoral injection of the indicated bacteria into mice bearing both CD20+ Namalwa and CD20− Karpas299 tumors (n = 8 mice per group). (C) Tracking of long-term Namalwa tumor growth coincident with the intratumoral administration of 2G9-AT-Salmonella (with or without HSV-TK), empty vector-Salmonella-HSV-TK, or PBS into mice bearing CD20+ Namalwa tumors, along with the intraperitoneal administration (twice daily) of either ganciclovir (100 mg/kg/day) or PBS. All data shown are taken from single experiments, which were representative of at least 2 separate experiments yielding similar results. (D) Actual cfus injected as determined after injection by limiting dilution assay of the injected bacterial solution, corresponding to the data presented in (C). (E) Tracking of Namalwa tumor growth after intravenous injection of 2G9-AT-HSV-TK (n = 7), 2G9-AT (n = 4), empty vector-AT-HSV-TK (n = 5), or PBS (n = 4) bacterial strains, along with intraperitoneal administration of ganciclovir. (F) Tracking of murine CD20+MCA203 fibrosarcoma tumors after intravenous administration following the same schedules or groupings as indicated in (E).
Human xenograft treatment via 2G9-Salmonella bearing the HSV-TK enzyme. (A) Intracellular 2G9-Salmonella and empty vector-Salmonella measured 24 hours after intratumoral injection into mice bearing both CD20+ and CD20− Karpas299 tumors. (B) Tracking of acute-phase tumor growth after a single intratumoral injection of the indicated bacteria into mice bearing both CD20+ Namalwa and CD20− Karpas299 tumors (n = 8 mice per group). (C) Tracking of long-term Namalwa tumor growth coincident with the intratumoral administration of 2G9-AT-Salmonella (with or without HSV-TK), empty vector-Salmonella-HSV-TK, or PBS into mice bearing CD20+ Namalwa tumors, along with the intraperitoneal administration (twice daily) of either ganciclovir (100 mg/kg/day) or PBS. All data shown are taken from single experiments, which were representative of at least 2 separate experiments yielding similar results. (D) Actual cfus injected as determined after injection by limiting dilution assay of the injected bacterial solution, corresponding to the data presented in (C). (E) Tracking of Namalwa tumor growth after intravenous injection of 2G9-AT-HSV-TK (n = 7), 2G9-AT (n = 4), empty vector-AT-HSV-TK (n = 5), or PBS (n = 4) bacterial strains, along with intraperitoneal administration of ganciclovir. (F) Tracking of murine CD20+MCA203 fibrosarcoma tumors after intravenous administration following the same schedules or groupings as indicated in (E).
The absence of adaptive immunity allowed us to therefore evaluate the usefulness of Salmonella to intracellularly target tumor cells for the delivery of a prodrug-converting enzyme. Thus, we generated 2G9-Salmonella expressing the HSV-TK enzyme (data not shown). This would allow us to deliver HSV-TK only into CD20-expressing cells, by means of the 2G9-Salmonella infection, thus sensitizing them to the prochemotherapeutic, ganciclovir, which is converted into a deoxyguanosine triphosphate analog that induces apoptosis on DNA incorporation.17 NOD/SCID animals bearing Namalwa tumors received 1 × 108 cfus of either 2G9-AT-HSV-TK, empty vector-AT-HSV-TK, 2G9-AT, or PBS intratumorally every 4 days with or without intraperitoneal ganciclovir injection [100 mg/kg/day].24 Only the 2G9-AT-HSV-TK, in combination with ganciclovir, delayed tumor growth or initiated regression (Figure 5C). Ganciclovir alone or combined with empty vector-HSV-TK-Salmonella failed to slow tumor growth. As seen in Figure 5C, average tumor dimensions were significantly smaller for the 2G9-AT-HSV-TK-ganciclovir combination therapy, compared with all other conditions tested (day 12 compared with empty vector-HSV-TK-ganciclovir [P = .02] or PBS-ganciclovir [P = .0001]). Likewise, the clinical response for this combination was vastly better with 2 of 5 animals exhibiting delayed tumor growth and 2 of 5 showing tumor reduction, 1 of which completely rejected the tumor. Remarkably, in more than 50 NOD/SCID tumor-bearing animals treated with PBS-ganciclovir alone, empty vector-AT-HSV-TK-ganciclovir, or 2G9-HSV-TK or empty vector-AT-HSV-TK alone, not a single animal exhibited delayed tumor growth or tumor regression. Actual cfus were verified to be identical among all tested conditions (Figure 5D). Although this result was quite promising, its usefulness is limited to only those tumors easily discovered and injected. Therefore, we sought to deliver the engineered Salmonella intravenously and track its effect on tumor growth. Strikingly, 2G9-AT-HSV-TK was quite successful in mediating significant reduction in tumor volume when coadministered with ganciclovir in 86% of the treated mice (Figure 5E). All other treatment regimens (2G9-AT-ganciclovir, empty vector-AT-HSV-TK-ganciclovir, or PBS-ganciclovir) failed to delay tumor progression. Similar results were obtained by intravenous administration into immune-competent mice bearing CD20+MCA203 tumors (Figure 5F). Again, 2G9-AT-HSV-TK-ganciclovir treatment induced tumor shrinkage in 71% and, remarkably, complete tumor ablation in 57% of the treated animals. Additionally, 2G9-AT-ganciclovir in this case achieved tumor reduction in 50% of treated animals before relapse occurred. Again, no animals receiving empty vector-AT-HSV-TK-ganciclovir or PBS-ganciclovir had significant delays in tumor growth. Therefore, the data indicate that we have created a novel therapeutic strategy allowing for the targeting and rejection of human tumors regardless of the presence of a functional adaptive immune response by using tumor-targeted Salmonella carrying a prodrug-converting enzyme with a systemically administered prochemotherapeutic agent.
Discussion
Survival of neoplastic cells despite constant immune monitoring is a key step that is acquired during tumorigenesis. The mechanisms comprising tumor immune evasion pose significant challenges to the application of immunotherapies. Indeed, it has proven difficult to boost preexisting anti-tumor immunity or generate it de novo in patients exhibiting widespread disturbances in the immune system. Therefore, new therapeutic approaches that circumvent immune escape are critical. The efficacy of Salmonella to target and eradicate growing tumors has been well documented.2,4,5 However, colonization and persistence of Salmonella within healthy tissues weaken its therapeutic efficacy and causes undesirable adverse effects.3,26 Therefore, it became necessary to develop novel methods to boost the tumor tropism of Salmonella.
This development was accomplished in our current study via the surface expression of a single-domain antibody vs a tumor-associated antigen (hCD20). Bacteria overexpressing cell surface 2G9 efficiently bound and entered CD20+ MCA203, CT26, and Namalwa cells, whereas Salmonella overexpressing GST-specific VHH failed to appreciably do so. Uptake of 2G9-Salmonella was dependent on the recognition of membrane-expressed hCD20, culminating in the remission and rejection of CD20+ tumors. Whereas others have previously used concomitant delivery of bispecific antibody and Salmonella mutants,29 or tumor-targeting VHHs30 to facilitate tumor rejection, our novel methodology uses viable bacteria engineered to express all moieties required for tumor destruction. Thus, we can create patient-specific “intelligent missiles” delivering a variety of cytotoxic payloads into targeted tumor cells, thus facilitating their specific destruction.
Accordingly, we found that the expression of VHH molecules almost completely ablated the generic cell infectivity of Salmonella in vitro and in vivo. Diminished nonspecific cell invasion strongly reduced the undesirable accumulation of Salmonella within mouse spleens and livers, thereby boosting their therapeutic usefulness. As has been proposed elsewhere,18 a fuller understanding of how bacteria colonize healthy and tumor tissues could give insight into how the tumors themselves grow and metastasize. Additionally, it would be interesting to assess whether enhanced tumor targeting and invasion may disfavor biofilm formation that could otherwise diminish anticancer activity of Salmonella.31
These findings represent a clear improvement toward the control of in vivo–delivered bacteria. The highly attenuated salmonellae currently approved for in vivo use, by necessity, lack bacterial-dependent cellular cytotoxicity.4 Rather, these vectors depend on the patient’s immune system to recognize and destroy infected cells. Indeed, we have recently shown that systemically administered Salmonella mediated an anti-tumor response via the DC upregulation of connexin43, thereby mediating GJ-dependent transfer of tumor antigens into recruited DCs, which, in turn, educate tumor-specific CD8+ T cells to reject tumors.8 Such a strategy may work for patients with a robust immune system, a condition often missing in advanced cancers. The ability to load tumor-targeted Salmonella with cytotoxic cargos may augment the immunostimulatory activity of Salmonella with a cytotoxic response specifically within tumor cells. Therefore, we engineered 2G9-Salmonella expressing the HSV-TK enzyme, which imparted ganciclovir sensitivity to infected cells. Accordingly, only CD20+ human xenografts cotreated with 2G9-Salmonella and ganciclovir underwent tumor reduction or rejection (Figure 5D-E). A similar system has previously been used to infect and visualize tumors via the HSV-TK–dependent incorporation of radionucleotides.32 The successful disruption of human tumor growth in NOD/SCID animals indicates that cytotoxicity occurs independently of adaptive immunity and may thus be a valuable tool to treat immunocompromised patients without safety concerns. Furthermore, our observation that antibody-targeted Salmonella can specifically enter lymphoma cells while being rapidly cleared from healthy tissues strongly suggests its usefulness for treatment of blood-borne cancers, a property hitherto unattributed to bacterial immunotherapies.
In conclusion, we have developed a novel methodology to generate cell/tumor-specific invasive Salmonella via the expression of VHH antibodies on their outer surface. Consequently, Salmonella entry occurs only into cells displaying the target antigen, abolishing generic bacterial cell entry both in vitro and in vivo. The effectiveness of targeted bacteria coexpressing HSV-TK, delivered either intravenously or intratumorally, indicates that the bacteria can be engineered to transport cytotoxic molecules into tumors and metastases, destroying them in the absence of adaptive immunity. Similarly, Salmonella can be loaded with radioisotopes,32 short hairpin RNAs,7,8 or tumor-exclusive immunogenic antigens33,34 to maximize tumor-specific killing.
Our work, therefore, represents a significant advancement toward the creation of safe, therapeutically efficacious Salmonella for the treatment of both detected and undetected tumors in humans. Furthermore, the ease with which specific VHHs can be isolated can streamline the creation of patient-specific Salmonella vectors via high-throughput screening for VHHs preferentially engaging patient tumors.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Drs Luigi Naldini, Vincenzo Russo, and Kenneth B. Marcu for sharing their plasmids and Dr Mario Colombo for the gift of the MCA203 and CT-26 cell lines.
M.R. is supported by grants from the European Commission (ERC, Dendroworld), the Italian Association for Cancer Research (AIRC), the Association for International Cancer Research (AICR) and the Ministry of Health (Ricerca Finalizzata).
Authorship
Contribution: P.E.M. performed and designed research, analyzed data and statistics, and performed manuscript preparation; A.P. collected data and analyzed data and statistics; A.M. performed experimental design and contributed vital new tools; A.d.M. performed research design and manuscript editing; and M.R. performed research design, data analysis, and manuscript editing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Maria Rescigno, Department of Experimental Oncology, European Institute of Oncology, Via Adamello 16, 20139 Milan, Italy; e-mail: maria.rescigno@ieo.eu.


![Figure 2. 2G9-Salmonella efficiently interacts with and invades CD20-expressing cells. (A) Representative immunofluorescence images of CD20-expressing CT26 cells infected with control AT-Salmonella (left panel), 2G9-Salmonella (middle panel), or GSTb1-Salmonella (right panel) at MOI 10, showing extracellular bacteria (Cy5-positive [blue]), intracellular bacteria (Cy3-Cy5-double positive [red]), and cytoplasm/phalloidin (FITC [green]) staining. Images shown are representative of 3 separate immunofluorescence infection experiments comprising at least 5 randomly selected fields of view per infection. (B) Quantification of total number of bound per infection (extracellular + intracellular), determined from at least 4 immunofluorescence images per infection, normalized per the cell count in field of view. (C) Quantification of invasiveness of bound bacteria (intracellular/[intracellular + extracellular]). (D) Total number of bacterial cfus used in (A) and (B), as determined by limiting dilution assay of the bacteria used for infection. All images shown were taken with a 40× objective.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/5/10.1182_blood-2012-12-474098/4/m_705f2.jpeg?Expires=1767720893&Signature=ictoyzI7AdVRGXgSU6duMqd42XPZzSrpdxnEAH8DHluttR-3daRpoXxsRAkxQ-bLnyygwFZUpuhKChzJRERWa6QOeoFUQjv7q~Ga4l-qgioyuKBEqDTlpIoO0bhbMJu6EUdgZDuNDvju6w4s8846EdAU8PLeQfczTpEc3xc6VCBbdH8syV7ZTSZNEcnAegvTPGcc~QUxnI7ng~JFKnkM8hIu-XG7YsM8fuGy-Q8LMPXfE0dfYeSsT6JcP73rmjANOYErRBjM9LXjpWOfYaocgMunRnZvei1WOeA6vu4jnpWGGVynmGLl4HbQ5M-uu9SV6yLuHNU0Wy-NsyknMZRXaA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. VHH-expressing Salmonella do not significantly invade CD20− cells. Representative immunofluorescence images of empty vector–transfected (A) MCA203 and (B) CT26 cells infected with control AT-Salmonella (left panels) or 2G9-Salmonella (right panels) at MOI 10, showing extracellular bacteria (Cy5-positive [blue]), intracellular bacteria (Cy3-Cy5-double positive [red]), and cytoplasm/phalloidin (FITC [green]) staining. Images shown are representatives of 3 separate immunofluorescence-infection experiments comprising at least 4 randomly selected fields of view per infection. (C) Quantification of total number of bound per infection (extracellular + intracellular), determined from 4 immunofluorescence images per infection, normalized per the cell count in field of view. (D) Quantification of invasiveness of bound bacteria (intracellular/[intracellular + extracellular]) shown in Figure 3C.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/5/10.1182_blood-2012-12-474098/4/m_705f3.jpeg?Expires=1767720893&Signature=Z8ILJt8llocMc8kRSg337B7RL2XAuuu8onQ8AHpRua5HyMiau-oqiKPij3G0YSft-OIj5vei~Mp40D2K7zSJ0BH~pfCFzmUcuppFcb3NxN91eRLCx1Y-zDrbTjrzCDoa~91Wg90AnUTi4EQBi9Bp-GzDv8C-Zu8nPfMg7fSjG3TN3eHTyTV72TiXjE8mTpHjyFm4UyWOTo9Xn8n97CAd29eMSZV-ESrqMmkTeRR2Bdgp7vx5Q0aewO8KQjIpoALp5h2asRnjFX-0t8vn80YKcQ6VFR1CHzUxlRRnjttEZJX9c2SdWMWSjlEfNTx2l9CvBOWIYnbfHScCBKlW99OxKg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 4. 2G9-Salmonella specifically invades and rejects CD20-positive tumors in vivo, increasing mouse survival duration. (A) Quantification of in vivo intracellular 2G9-Salmonella vs GSTb1-Salmonella within CD20+ and CD20− MCA203 tumors, 24 hours after intravenous delivery (n = 4 per group). (B) In vivo, intracellular organ biodistribution (liver, spleen, and tumor) of unmodified AT-Salmonella vs 2G9-Salmonella 24 hours after intratumoral delivery as assessed by gentamicin protection assay. n.d.= not detectable. (C-D) Tracking of acute-phase tumor growth after a single intratumoral injection of the indicated bacteria into mice bearing both CD20+ and CD20− MCA203 tumors in opposite flanks (n = 5 mice per group [PBS and empty vector-AT-Salmonella] or 8 mice per group [2G9-AT]). (E) Kaplan-Meier curves of mice bearing either CD20+ or CD20− MCA203 tumors injected twice with 2G9-Salmonella or empty vector-Salmonella (n = 6 mice per group). (F) Actual cfus injected as determined after injection by limiting dilution assay of the injected bacterial solution, bars 1 and 2 corresponding to the data presented in (A) and bars 3 and 4 corresponding to the data shown in (C) and (D). All data shown are from representative experiments, which were repeated 3 times, yielding similar results.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/5/10.1182_blood-2012-12-474098/4/m_705f4.jpeg?Expires=1767720893&Signature=MR02NwSJxb6Up6TBZrDGprEzYC4SdcKvAEkyRXw3BEj9lm3L4Q~7sjWZMhHUCUOqRpzrDbidh4aMW51dmQgDNas8S~aj8lnweSoWsiMOw3vIgv~obfhtOiWXc54bqw9QEdS4uDo9e6lNxd6bchTYyq00tucl-uOPN4VXvfGRwHCm237uEfmXTdFqap8MkZ-YiLHcZWnUtI6mtluIxFru3W5Wfx3guJyJ3KDoQN4JykDSDkceAr6aTpsSdeii1z4hPQIP4lOz5Ng9O6HAEyEUS5Gmbq05wmxlnQDDk-I1MKPVWoE6NnuQo0lDWbLdB2BFWNlTHSDEChGXCNdM9IqTcA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal