Key Points
B7-H6 transcripts, B7-H6 cell-surface expression, and sB7-H6 can be induced in inflammatory conditions in vitro and in vivo.
B7-H6 is expressed on proinflammatory CD14+CD16+ monocytes in sepsis conditions and is linked to an increased mortality.
Abstract
B7-H6, a member of the B7 family of immunoreceptors, is as a cell-surface ligand for the NKp30-activating receptor expressed on natural killer cells. B7-H6 is not detected in normal human tissues at steady state but is expressed on tumor cells. However, whether B7-H6 can be expressed in other conditions remains unknown. We analyzed here the pathways that lead to the expression of B7-H6 in nontransformed cells. In vitro, B7-H6 was induced at the surface of CD14+CD16+ proinflammatory monocytes and neutrophils upon stimulation by ligands of Toll-like receptors or proinflammatory cytokines such as interleukin-1β and tumor necrosis factor α. In these conditions, a soluble form of B7-H6 (sB7-H6) was also produced by activated monocytes and neutrophils. In vivo, B7-H6 was expressed on circulating proinflammatory CD14+CD16+ monocytes in a group of patients in sepsis conditions, and was linked to an increased mortality. sB7-H6 was selectively detected in the sera of patients with gram-negative sepsis and was associated with membrane vesicles that co-sedimented with the exosomal fraction. These findings reveal that B7-H6 is not only implicated in tumor immunosurveillance but also participates in the inflammatory response in infectious conditions. This trial was registered at www.clinicaltrials.gov as #NTC00699868.
Introduction
Natural killer (NK) cells are cytolytic and cytokine-producing lymphocytes that can recognize a variety of cells in distress.1 Cells undergoing microbial infection, tumor transformation, as well as chemical or physical insults become NK-cell targets when they express stress-induced surface molecules that interact with activating NK cell-surface receptors. The recognition of NKG2D ligands expressed by target cells via NKG2D expressed on NK cells represents a prototypical illustration of this stress-induced self-mode of innate immune recognition.2
NK cells express at their surface a group of activating receptors that are immunoglobulin (Ig)-like superfamily members and are referred as to natural cytotoxicity receptors (NCRs). NCRs include NKp46 (NCR1, CD335), NKp44 (NCR2, CD336), and NKp30 (NCR3, CD337).3 Although these molecules have no homology, they have been grouped as NCRs based on the similarities in their expression profile, their oligomeric structure, and their function.4 The characterization of the NCR ligands is still incomplete. Although many data support a central role of NCR in tumor surveillance,5-8 the first NCR ligands to be identified were viral structures, in particular the influenza hemagglutinin for NKp469 and the human cytomegalovirus pp65 tegument protein for NKp30.10 Later, the HLA-B–associated transcript 3 (BAT3) protein was shown to bind and trigger NKp30.11 This nuclear protein now referred to as BAG6 (http://www.genenames.org/data/hgnc_data.php?hgnc_id=13919) is ectopically found at the plasma membrane upon stress but absent from tumor cells susceptible to NK-cell lysis. We identified B7-H6 (NCR3LG1, http://www.genenames.org/data/hgnc_data.php?hgnc_id=42400) as a ligand of NKp30.12,13 B7-H6 transcripts have not been detected in normal adult tissues. However, B7-H6 is present on a broad panel of hematopoietic and nonhematopoietic tumor cells including lymphoma, leukemia, melanoma, and carcinoma as well as on primary tumor blood cells.12 The pattern of B7-H6 expression, which appears so far to be limited to tumor cells, is thus another example of stress-induced self-recognition by NK cells. In the case of NKG2D ligands, several mechanisms have been shown to regulate their expression. For example, genotoxic stress was shown to upregulate NKG2D ligands on cell lines, whereas interfering with the DNA damage pathway in tumor cells inhibited their constitutive expression.2 In contrast to NKG2D ligands, the regulation of B7-H6 expression is unknown. We studied here the mechanisms that govern the induction of B7-H6 on primary cells as well as its consequences in vitro and in vivo.
Methods
A detailed description of all experimental methods can be found in the supplemental Methods (available on the Blood website).
Peripheral blood mononuclear cells (PBMCs) were isolated from healthy volunteer donors using Ficoll-Paque Plus (GE Healtcare) density centrifugation. Human monocytes were purified with CD14 microbeads (Miltenyi Biotec) according to the manufacturer’s instructions. Cell purity was 90% to 98%. Human neutrophils were isolated from peripheral blood of healthy donors using a dextran-Ficoll method. Human NK cells were purified with a human NK cell isolation II kit (Miltenyi Biotec) according to the manufacturer’s instructions. Cells were cultured in complete RPMI 1640 medium (Gibco; Life Technologies) supplemented with 100 U/mL penicillin/streptomycin (Gibco; Life Technologies), 1mM sodium pyruvate (Gibco; Life Technologies), 10% heat-inactivated fetal bovine serum (FBS) (Lonza). Human PBMCs (3 × 106 cells), neutrophils (2 × 106 cells), or monocytes (5 × 105 cells) were seeded into 24-well plates (BD). Cells were then incubated at 37°C with 5% CO2 in the presence or absence of the following stimuli: 300 ng/mL synthetic bacterial lipoprotein (Pam3CSK4), 10 µg/mL zymosan, 108 cells/mL heat-killed Acholeplasma laidlawii (HKAL), Listeria monocytogenes (HKLM), Helicobacter pylori (HKHP) or Staphylococcus aureus (HKSA), 500 ng/mL synthetic diacylated lipoprotein (FSL-1), 1 µg/mL ultra-pure lipopolysaccharide (LPS) from Escherichia coli, 1 µg/mL flagellin from Staphylococcus typhimurium, 20 µg/mL imiquimod (R837), 5 µg/mL single-stranded RNA40, 20 µg/mL oligodeoxynucleotides (ODNs) CpG type A, 100 µg/mL poly(I:C), 100 ng/mL tumor necrosis factor α (TNFα), or 1 ng/mL interleukin-1β (IL-1β). All reagents were purchased from Invivogen except for IL-1β that was purchased from PeproTech. The study trial (clincaltrials.gov NTC00699868) was approved by the Sud-Méditerranée V Ethics Committee (Comité de Protection des Personnes). Written informed consent was obtained from all patients or their proxies in accordance with the Declaration of Helsinki.
Results
In vitro induction of B7-H6 transcripts in primary blood cells
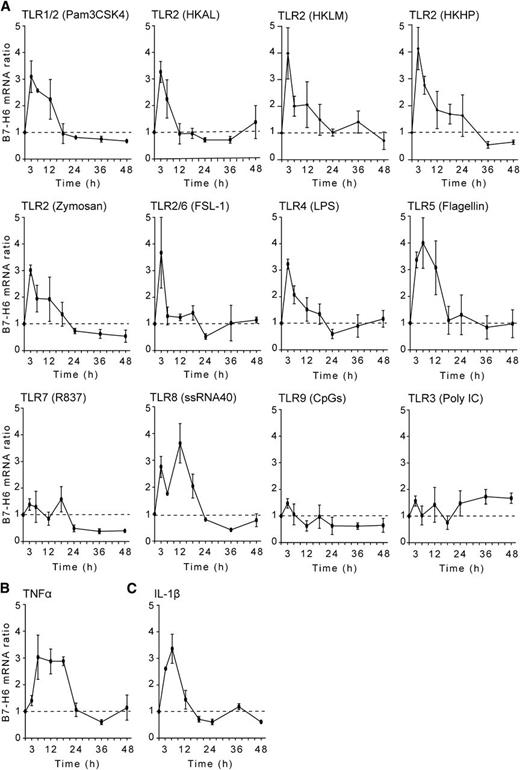
We set up an in vitro screening test to investigate whether B7-H6 could be induced on primary blood cells. In this assay, PBMCs were treated with a variety of agents, and messenger RNA (mRNA) was extracted to quantify B7-H6 transcripts using quantitative real-time–polymerase chain reaction. In contrast to reports showing that DNA-damaging agents (such as irradiation, cisplatine, etoposide, or fluorouracile) and proteasome inhibitors (such as epoxomicin or MG132) can induce the expression of NKG2D ligands,2 these treatments had no substantial effect on B7-H6 expression in our protocols (data not shown). However, stimulations of PBMCs with a panel of Toll-like receptor (TLR) agonists induced B7-H6 transcripts. TLR ligands that are recognized via TLR2 (such as Pam3CSK4, HKAL as an example of cell-wall–less bacteria, HKLM as an example of gram-positive bacteria, and HKHP as an example of gram-negative bacteria, zymosan, and FSL-1), or via TLR4 (such as LPS), or via TLR5 (such as flagellin) induced a fast expression of B7-H6 mRNA (Figure 1A). In contrast, the stimulation of PBMCs via endosomal TLRs, such as TLR7 (R837), TLR9 (CpGs), and TLR3 (Poly IC) had very limited impact on B7-H6 mRNA expression. A noticeable exception was the stimulation of primary cells by single-stranded RNA that triggers a TLR8-dependent pathway and induced B7-H6 transcripts (Figure 1A). As a control, all TLR stimulation induced IL-6 transcripts (supplemental Figure 1). It is noteworthy that the proinflammatory cytokines TNFα and IL-1β also induced B7-H6 transcripts (Figure 1B-C). Overall, the kinetics of B7-H6 mRNA induction upon TLR and cytokine secretion were quite similar, peaking early between 3 and 12 hours, and returning fast to baseline within 24 hours poststimulation. Thus, B7-H6 is one of the early genes that are induced in primary blood human cells upon inflammatory and microbial stimulations in vitro.
In vitro induction of B7-H6 transcripts.B7-H6 mRNA is upregulated in PBMCs during activation by (A) TLR ligands, (B) TNFα, or (C) IL-1β. Control cells were treated with medium only. Data show fold induction of B7-H6 mRNA in treated cells as compared with untreated control cells at the indicated time points. Data correspond to a pool of 3 independent experiments.
In vitro induction of B7-H6 transcripts.B7-H6 mRNA is upregulated in PBMCs during activation by (A) TLR ligands, (B) TNFα, or (C) IL-1β. Control cells were treated with medium only. Data show fold induction of B7-H6 mRNA in treated cells as compared with untreated control cells at the indicated time points. Data correspond to a pool of 3 independent experiments.
In vitro induction of cell-surface B7-H6
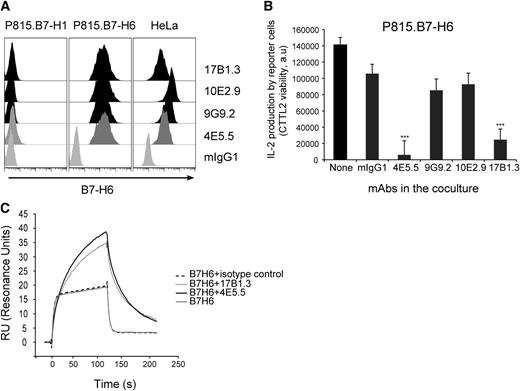
To further investigate the regulation of B7-H6 expression, we generated a panel of mouse monoclonal antibodies (mAbs) directed against human B7-H6. Four mAbs were selected on the basis of their selective reactivity with P815.B7-H6, but not with P815.B7-H1 stable transfectants (Figure 2A). As expected, they also reacted with human tumor cell lines constitutively expressing B7-H6 such as HeLa cells (Figure 2A). Two of these mAbs, 4E5.5 and 17B1.3, were blocking the activation of NKp30+ reporter cells (DOMsp30) induced by P815.B7-H6 (Figure 2B). Despite their blocking activity, surface plasmon resonance (SPR) analysis showed that they did not inhibit the direct interaction of B7-H6 with NKp30 (Figure 2C). Indeed, the complexes formed between B7-H6 and anti-B7-H6 mAbs not only bound to immobilized soluble recombinant NKp30, but also showed enhanced binding and better stability, as compared with B7-H6 alone (Figure 2C). These data contrast with the inhibition of the interaction between NKp30 and B7-H6 induced by 2 blocking anti-NKp30 mAbs (Az20 and 1849; supplemental Figure 2), and prompts further investigation on the mechanisms by which 4E5.5 and 17B1.3 mAbs exert their blocking activity.
Characterization of anti-B7-H6 mAbs. (A) P815.B7-H1, P815.B7-H6, or HeLa cells were stained with 4E5.5, 9G9.2, 10E2.9, or 17B1.3 anti–B7-H6 mAbs and analyzed by flow cytometry. mIgG1 was used as isotype control. Graphs are representative of at least 3 experiments. (B) NKp30+ DOMsp30 reporter cells were cocultured with P815.B7-H6 cells in presence or absence of anti–B7-H6 mAbs. DOMsp30 cell activation was determined by evaluating IL-2 production in the coculture supernatant in a standard CTLL-2 survival assay. Data are representative of 3 independent experiments. ***P < .001. (C) SPR analysis: Superimposed sensorgrams showing the injections onto NKp30 chip of soluble recombinant B7-H6 alone or preincubated with anti–B7-H6 mAbs. Sensorgrams were normalized in the y-axis and aligned in the x-axis at the end of injection. Sensorgrams are representative of 2 independent experiments. a.u., arbitrary unit; mIgG1, mouse IgG1.
Characterization of anti-B7-H6 mAbs. (A) P815.B7-H1, P815.B7-H6, or HeLa cells were stained with 4E5.5, 9G9.2, 10E2.9, or 17B1.3 anti–B7-H6 mAbs and analyzed by flow cytometry. mIgG1 was used as isotype control. Graphs are representative of at least 3 experiments. (B) NKp30+ DOMsp30 reporter cells were cocultured with P815.B7-H6 cells in presence or absence of anti–B7-H6 mAbs. DOMsp30 cell activation was determined by evaluating IL-2 production in the coculture supernatant in a standard CTLL-2 survival assay. Data are representative of 3 independent experiments. ***P < .001. (C) SPR analysis: Superimposed sensorgrams showing the injections onto NKp30 chip of soluble recombinant B7-H6 alone or preincubated with anti–B7-H6 mAbs. Sensorgrams were normalized in the y-axis and aligned in the x-axis at the end of injection. Sensorgrams are representative of 2 independent experiments. a.u., arbitrary unit; mIgG1, mouse IgG1.
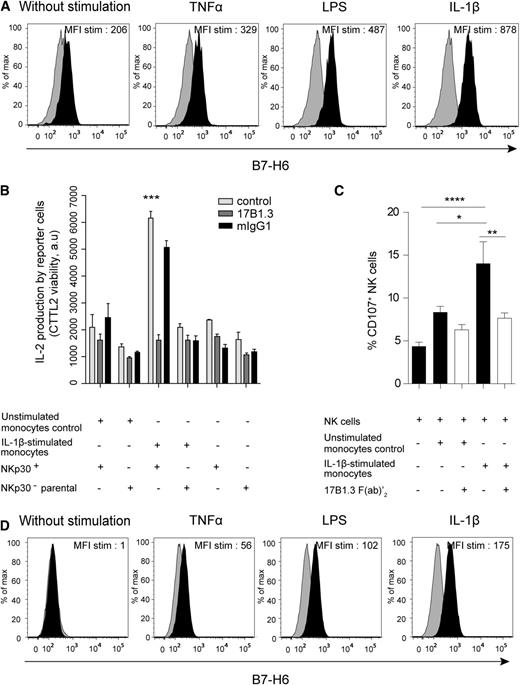
The availability of these anti-B7-H6 mAbs allowed us to investigate whether the in vitro inducers of B7-H6 transcripts also led to the cell-surface expression of the protein and on which cell type. PBMCs were therefore stimulated with TLR agonists, IL-1β or TNFα, for 48 hours, and the cell-surface expression of B7-H6 was analyzed by flow cytometry. At steady state, no cell-surface expression of B7-H6 on PBMCs could be detected (Figure 3A), consistent with previous findings.12 At 48 hours, B7-H6 was expressed on the surface of CD45+CD14+CD19−CD3− monocytes upon TNFα, LPS, flagellin, and IL-1β stimulation (Figure 3B), consistent with the induction of B7-H6 transcripts in response to these stimulations. The induction of B7-H6 was specific of monocytes, as all others cells from PBMCs remain negative for B7-H6 expression upon in vitro stimulation with TLR ligands or cytokines (data not shown). A low but reproducible induction of B7-H6 protein was seen on monocytes cultured for 48 hours on plastic dishes without any further stimulation (Figure 3B). Poly IC did not lead to the cell-surface expression of B7-H6 on monocytes (Figure 3C), consistent with our transcript expression data. Despite a rapid decline in B7-H6 transcripts (Figure 1A), the cell-surface expression of the B7-H6 protein on monocytes was stable up to 48 hours (Figure 3C). The decrease in monocyte viability upon a longer period of in vitro culture prevented us from further analyzing the kinetics of B7-H6 cell-surface expression. CD14+ monocytes were also purified from PBMCs and stimulated in vitro for 48 hours with TLR ligands or proinflammatory cytokines. Under these conditions, purified monocytes became B7-H6+, indicating that the induction of B7-H6 on monocytes was the consequence of direct effect of inflammatory or microbial stimulations on these cells (Figure 4A). Even if freshly purified monocytes contain both CD14+CD16− and CD14+CD16+ subsets, upon in vitro culture in the presence or absence of additional stimuli, all monocytes acquired the CD14+CD16+ proinflammatory type (supplemental Figure 3).14 Although the density of B7-H6 molecules expressed at the surface of monocytes was moderate, it was sufficient to trigger the activation of the NKp30+ DOMsp30 reporter cells. This activation was blocked by the 17B1.3 anti–B7-H6 blocking mAbs (Figure 4B). As a control, no stimulation of the NKp30− parental DO11.10 cells was induced by activated monocytes (Figure 4B). Importantly, IL-1β–stimulated B7-H6+ monocytes triggered autologous NK cells, and this activation was in an B7-H6–dependent manner, as shown by the inhibitory effect of anti–B7-H6 F(ab′)2 fragments (Figure 4C).
In vitro induction of B7-H6 cell-surface expression on monocytes. Flow cytometric analysis of CD45+CD14+CD19−CD3− monocytes, gated from freshly isolated PBMCs, (A) left untreated or (B) after stimulation for 48 hours with TNFα, LPS, flagellin, or IL-1β. B7-H6 expression was analyzed by flow cytometry with directly conjugated 17B1.3 anti–B7-H6 mAbs (black histograms) or mIgG1 isotype control (gray histograms). Data are representative of at least 3 independent experiments. MFI stim represents the value of the MFI obtained with anti–B7-H6 mAbs minus the MFI obtained with mIgG1 isotype control. (C) PBMCs were left untreated or treated with Poly IC, TNFα, LPS, flagellin, or IL-1β at indicated time points. B7-H6 cell-surface expression was assessed by flow cytometry and fold change in B7-H6 expression was quantified by dividing the MFI of treated samples by that of untreated cells at each time points. Data correspond to a pool of at least 3 independent experiments. Statistical analyses were performed using the 2-way ANOVA test with Bonferroni correction. *P < .05; **P < .01; and ***P < .001. ANOVA, analysis of variance.
In vitro induction of B7-H6 cell-surface expression on monocytes. Flow cytometric analysis of CD45+CD14+CD19−CD3− monocytes, gated from freshly isolated PBMCs, (A) left untreated or (B) after stimulation for 48 hours with TNFα, LPS, flagellin, or IL-1β. B7-H6 expression was analyzed by flow cytometry with directly conjugated 17B1.3 anti–B7-H6 mAbs (black histograms) or mIgG1 isotype control (gray histograms). Data are representative of at least 3 independent experiments. MFI stim represents the value of the MFI obtained with anti–B7-H6 mAbs minus the MFI obtained with mIgG1 isotype control. (C) PBMCs were left untreated or treated with Poly IC, TNFα, LPS, flagellin, or IL-1β at indicated time points. B7-H6 cell-surface expression was assessed by flow cytometry and fold change in B7-H6 expression was quantified by dividing the MFI of treated samples by that of untreated cells at each time points. Data correspond to a pool of at least 3 independent experiments. Statistical analyses were performed using the 2-way ANOVA test with Bonferroni correction. *P < .05; **P < .01; and ***P < .001. ANOVA, analysis of variance.
In vitro induction of B7-H6 cell-surface expression on purified myeloid cells and NKp30-dependent cell activation induced by B7-H6+ monocytes. (A) Flow cytometric analysis of B7-H6 cell-surface expression on freshly isolated monocytes stimulated for 48 hours with TNFα, LPS, or IL-1β. B7-H6 expression was analyzed by flow cytometry with directly conjugated 17B1.3 anti–B7-H6 mAbs (black histograms) or mIgG1 isotype control (gray histograms). Data are representative of at least 3 independent experiments. MFI stim represents the value of the MFI obtained with anti–B7-H6 mAbs minus the MFI obtained with mIgG1 isotype control. (B) NKp30+ DOMsp30 reporter cells (NKp30+) or NKp30− DO11.10 control cells (parental cells) were cocultured with unstimulated or IL-1β–stimulated monocytes in the presence of anti–B7-H6 mAbs (17B1.3) or mIgG1 isotype control. DOMsp30 cell activation was determined by evaluating IL-2 production in the coculture supernatant in a standard CTLL-2 survival assay. Data are representative of 3 independent experiments. ***P < .001. (C) Autologous NK cells were cocultured with unstimulated or IL-1β–stimulated monocytes (E:T, 1:4) in the presence of anti–B7-H6 (17B1.3) F(ab′)2 fragments. NK-cell activation was determined by evaluating the percentage of CD107-positive NK cells. Data correspond to a pool of 6 independent experiments. *P < .05; **P < .01; and ****P < .0001. (D) Flow cytometric analysis of B7-H6 expression on freshly isolated CD24+CD14−HLA−DR− neutrophils left untreated or stimulated with TNFα, LPS, or IL-1β for 24 hours. B7-H6 expression was analyzed by flow cytometry with F(ab)′2 fragments of the B7-H6–specific 17B1.3 mAbs followed by APC-conjugated anti-mouse IgG (black histograms) or with APC-conjugated anti-mouse IgG alone (control gray histograms). Data are representative of at least 3 independent experiments. MFI stim represent the value of the MFI obtained with anti-B7-H6 F(ab)′2 minus the MFI obtained with the control. E:T, effector to target ratio.
In vitro induction of B7-H6 cell-surface expression on purified myeloid cells and NKp30-dependent cell activation induced by B7-H6+ monocytes. (A) Flow cytometric analysis of B7-H6 cell-surface expression on freshly isolated monocytes stimulated for 48 hours with TNFα, LPS, or IL-1β. B7-H6 expression was analyzed by flow cytometry with directly conjugated 17B1.3 anti–B7-H6 mAbs (black histograms) or mIgG1 isotype control (gray histograms). Data are representative of at least 3 independent experiments. MFI stim represents the value of the MFI obtained with anti–B7-H6 mAbs minus the MFI obtained with mIgG1 isotype control. (B) NKp30+ DOMsp30 reporter cells (NKp30+) or NKp30− DO11.10 control cells (parental cells) were cocultured with unstimulated or IL-1β–stimulated monocytes in the presence of anti–B7-H6 mAbs (17B1.3) or mIgG1 isotype control. DOMsp30 cell activation was determined by evaluating IL-2 production in the coculture supernatant in a standard CTLL-2 survival assay. Data are representative of 3 independent experiments. ***P < .001. (C) Autologous NK cells were cocultured with unstimulated or IL-1β–stimulated monocytes (E:T, 1:4) in the presence of anti–B7-H6 (17B1.3) F(ab′)2 fragments. NK-cell activation was determined by evaluating the percentage of CD107-positive NK cells. Data correspond to a pool of 6 independent experiments. *P < .05; **P < .01; and ****P < .0001. (D) Flow cytometric analysis of B7-H6 expression on freshly isolated CD24+CD14−HLA−DR− neutrophils left untreated or stimulated with TNFα, LPS, or IL-1β for 24 hours. B7-H6 expression was analyzed by flow cytometry with F(ab)′2 fragments of the B7-H6–specific 17B1.3 mAbs followed by APC-conjugated anti-mouse IgG (black histograms) or with APC-conjugated anti-mouse IgG alone (control gray histograms). Data are representative of at least 3 independent experiments. MFI stim represent the value of the MFI obtained with anti-B7-H6 F(ab)′2 minus the MFI obtained with the control. E:T, effector to target ratio.
In humans, TLR2 is expressed by monocytes and neutrophils, TLR3 is expressed by BDCA3+ conventional dendritic cells (DCs), TLR5 is expressed by monocytes, neutrophils, and conventional DCs, TLR7 is expressed by monocytes and plasmacytoid DCs, and TLR8 is expressed by neutrophils and TLR9 by plasmacytoid DCs.15 The pattern of TLR expression was thus consistent with the selectivity of B7-H6 induction on monocytes, but also prompted us to examine whether neutrophils could express surface B7-H6 upon stimulation. Whereas unstimulated neutrophils do not express cell-surface B7-H6, treatment of neutrophils with TNFα, LPS, IL-1β (Figure 4D), HKSA, and HKHP (supplemental Figure 4) leads to a weak but reproducible induction of cell-surface B7-H6. As for monocytes, the effect of IL-1β was direct as it occurred on purified neutrophils. Anti–B7-H6 F(ab)′2 fragments were used to ascertain the specificity of anti–B7-H6 mAbs detection on these FcR+ myeloid cells (Figure 4D, supplemental Figure 4), as on monocytes (data not shown).
In vitro induction of soluble B7-H6
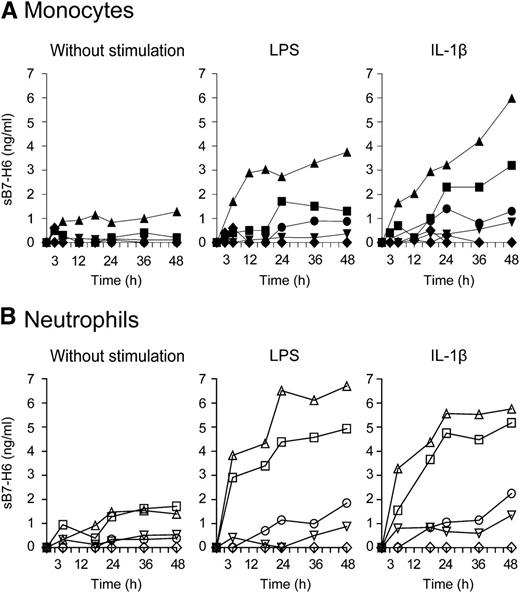
The availability of a panel of anti–B7-H6 mAbs allowed us to identify that 2 of them recognize nonoverlapping B7-H6 epitopes, rendering it possible to set-up a sandwich enzyme-linked immunosorbent assay (ELISA) against B7-H6 (supplemental Figure 5). We then investigated whether soluble forms of B7-H6 could be produced in vitro. Purified monocytes were activated for 48 hours with LPS or IL-1β and soluble B7-H6 (sB7-H6) was measured in cell supernatants by ELISA. No sB7-H6 was detected in the supernatant of monocytes in absence of stimulation. In contrast, sB7-H6 could be detected in the supernatant of LPS- and IL-1β–stimulated monocytes isolated from a group of healthy individuals (4 of 9) (Figure 5A). Similarly, neutrophils prepared from another group of healthy individuals and stimulated with LPS or IL-1β also produced sB7-H6 (4 of 6) (Figure 5B). HKHP was able to induce sB7-H6 from monocytes or neutrophils in vitro in some individuals, although stimulated monocytes and neutrophils from all healthy individuals expressed B7-H6 at the surface (supplemental Figure 4). The mechanisms underlying the heterogeneity between cells from healthy individuals that produce (or not) detectable amounts of sB7-H6 in vitro is still unclear. Irrespective of this issue that needs further investigation, these results reveal that a soluble form of B7-H6 can be produced by neutrophils and monocytes in response to inflammatory and microbial stimuli.
In vitro induction of sB7-H6 from human monocytes and neutrophils. (A) Freshly isolated monocytes or (B) freshly isolated neutrophils were treated with LPS or IL-1β for the indicated time points. Concentrations of sB7-H6 were measured in the cell supernatant by ELISA. Each symbol represents 1 individual donor. Graph represents 9 healthy individuals donors for monocytes and 6 healthy individuals donors for granulocytes.
In vitro induction of sB7-H6 from human monocytes and neutrophils. (A) Freshly isolated monocytes or (B) freshly isolated neutrophils were treated with LPS or IL-1β for the indicated time points. Concentrations of sB7-H6 were measured in the cell supernatant by ELISA. Each symbol represents 1 individual donor. Graph represents 9 healthy individuals donors for monocytes and 6 healthy individuals donors for granulocytes.
In vivo expression of B7-H6 during sepsis
The in vitro induction of B7-H6 by TLR ligands and proinflammatory cytokines prompted us to analyze the expression pattern of B7-H6 in inflammatory and infectious conditions in vivo. To address this point, PBMCs were obtained from a collection of peripheral blood samples prepared from a cohort of 39 patients presenting a systemic inflammatory response syndrome (SIRS) and admitted at the medical intensive care unit (ICU) (supplemental Table 1). Cells were harvested at the day of SIRS diagnosis (day 1) and kept frozen. Patients were divided into 2 categories: 27 patients with sepsis (sepsis group) and 12 patients with SIRS in absence of detectable microbial infection (SIRS group). Samples from 32 patients of 39 could be thawed successfully and PBMCs were analyzed for the cell-surface expression of B7-H6 on T cells (CD3+CD19−CD14−), B cells (CD3−CD19+CD14−), DCs (CD3−CD19−CD14−DR+), and monocytes (CD3−CD19−CD14+). B7-H6 expression was considered positive for samples for which the mean fluorescence intensity (MFI) ratio was strictly above 2. Using these criteria, the cell-surface expression of B7-H6 was detected in PBMCs of 15 patients (Figure 6A, supplemental Table 1). Remarkably, the cell-surface expression of B7-H6 was restricted to patients presenting sepsis (Figure 6). Moreover, this expression was selectively observed on CD14+CD16+ proinflammatory-type monocytes (Figure 6A), consistent with our in vitro data (Figure 4A, supplemental Figure 3). As expected, no cell-surface expression of B7-H6 was detected on monocytes from healthy control individuals (Figure 6C). The absence of neutrophils in frozen PBMC preparations prevented us from analyzing whether surface B7-H6 could be detected on these cells. sB7-H6 was also detected in the serum of 10 of 39 patients and restricted to the sepsis group (Figure 6D). No sB7-H6 was detected in sera obtained from other SIRS patients or healthy individuals (Figure 6D). Overall, 65% of patients with sepsis had circulating B7-H6+CD14+CD16+ proinflammatory-type monocytes, when no B7-H6+ PBMCs could be detected in other SIRS patients or healthy individuals. No difference was observed between sepsis patients presenting gram-negative infections (Achromobacter xylosoxidans, E coli, Enterobacter aerogenes, Klebsiella pneumoniae, and Pseudomonas aeruginosa) or gram-positive infections (S aureus, Streptococcus pneumoniae, and Streptococcus mitis). In contrast, serum sB7-H6 was detected in 37% of sepsis patients, and restricted to patients with gram-negative infection (Figure 6E, supplemental Table 2). Consistent with these results, gram-negative bacteria (eg, HKHP) are much more prone than gram-positive bacteria (eg, HKSA) to induce sB7-H6 in vitro (supplemental Figure 4). Most gram-negative sepsis patients who had detectable levels of serum sB7-H6 at day 1 also had circulating B7-H6+CD14+CD16+ proinflammatory monocytes on the same day (supplemental Table 1).
In vivo expression of B7-H6 on inflammatory monocytes during sepsis and association of B7-H6 and sepsis conditions. Flow cytometric analysis of B7-H6 expression on CD45+CD14+CD19−CD3− monocytes, gated from PBMCs of (A) sepsis patients, (B) SIRS patients, and (C) healthy donors. Cells are stained with CD14, CD16 mAbs, and directly conjugated 17B1.3 anti–B7-H6 mAbs (black histograms) or mIgG1 isotype control (gray histograms). Data are representative of 15 sepsis patients (23 patients in total), 9 SIRS patients, and 14 healthy donors. (D) Concentrations of sB7-H6 in the sera of sepsis patients, SIRS patients, or healthy donors were measured by ELISA. Statistical analyses were performed using the Student t test. (E) Sepsis patients were stratified according to the nature of their sepsis (induced by Gram+LPS− or Gram−LPS+ bacteria), and concentrations of sB7-H6 were measured by ELISA. Statistical analyses were performed using the Student t test. (F) Kaplan-Meier curves of survival probability obtained by segregating the whole cohort of ICU patients into 2 groups according to the expression of B7-H6 cell-surface expression. The number of the specimen is indicated on the graph. Statistical analyses were performed using the Pearson X2 test. mB7-H6+, B7-H6–positive cells; mB7-H6−, B7-H6–negative cells.
In vivo expression of B7-H6 on inflammatory monocytes during sepsis and association of B7-H6 and sepsis conditions. Flow cytometric analysis of B7-H6 expression on CD45+CD14+CD19−CD3− monocytes, gated from PBMCs of (A) sepsis patients, (B) SIRS patients, and (C) healthy donors. Cells are stained with CD14, CD16 mAbs, and directly conjugated 17B1.3 anti–B7-H6 mAbs (black histograms) or mIgG1 isotype control (gray histograms). Data are representative of 15 sepsis patients (23 patients in total), 9 SIRS patients, and 14 healthy donors. (D) Concentrations of sB7-H6 in the sera of sepsis patients, SIRS patients, or healthy donors were measured by ELISA. Statistical analyses were performed using the Student t test. (E) Sepsis patients were stratified according to the nature of their sepsis (induced by Gram+LPS− or Gram−LPS+ bacteria), and concentrations of sB7-H6 were measured by ELISA. Statistical analyses were performed using the Student t test. (F) Kaplan-Meier curves of survival probability obtained by segregating the whole cohort of ICU patients into 2 groups according to the expression of B7-H6 cell-surface expression. The number of the specimen is indicated on the graph. Statistical analyses were performed using the Pearson X2 test. mB7-H6+, B7-H6–positive cells; mB7-H6−, B7-H6–negative cells.
Despite the small size of our cohort of patients, we analyzed the potential association of B7-H6 expression and the clinical outcome of ICU patients. The overall day 30 mortality in ICU patients was 23.1%. We observed a higher mortality in sepsis patients with membrane B7-H6 (mB7-H6+) expression as compared with patients lacking mB7-H6+ cells (mB7-H6−) (Figure 6F). In particular, no death was observed when mB7-H6 expression was negative in patients with gram-negative sepsis, as compared with 44% death in patients expressing mB7-H6.
Analysis of sB7-H6
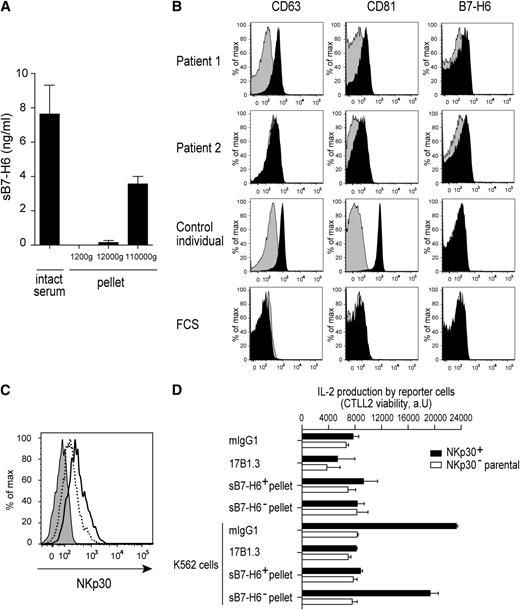
We took advantage of the substantial quantities of sB7-H6 present in patient sera to document the nature of sB7-H6. In a first series of experiments, patient sera were subjected to 3 rounds of centrifugations. The pellets obtained at each round were analyzed by ELISA for the presence of sB7-H6. sB7-H6 was associated with the material that sedimented at 110 000g, suggesting that sB7-H6 was included in membrane vesicles that were present in the serum, such as exosomes (Figure 7A). To further dissect this point, sB7-H6+ patient sera were analyzed for the presence of B7-H6+ membrane vesicles using sequential low-speed centrifugations and high-speed centrifugations followed by a 0.22μM filtration as described previously.16 After incubation of the filtered pellets with latex beads, we detected that sB7-H6+ patient sera contained filtered pellet material that was reactive with CD63, CD81, and B7-H6 mAbs (Figure 7B). CD63 and CD81 are classically considered as markers of exosomes,16 suggesting that B7-H6 can be associated with these membrane vesicles in the serum of sepsis patients. Incubation of primary NK cells with the sB7-H6+ pellets that sedimented with the exosomal fraction impaired the staining of NK cells with anti-NKp30 mAbs, whereas sB7-H6− pellets had no effect (Figure 7C). In addition, sB7-H6+ pellets also inhibited the NKp30-dependent activation of reporter cells induced by B7-H6+ K562 cells as efficiently as the blocking 17B1.3 anti-B7-H6 mAbs (Figure 7D). Interestingly, recombinant sB7-H6 had no effect on NKp30 cell-surface expression and on NKp30-dependent cell activation over a large range of concentrations (data not shown). The available anti-NKp30 mAbs used to stain NK cells were blocking anti–NKp30 reagent,7 making it impossible to formally show that sB7-H6+ pellets downregulated NKp30 cell-surface expression or merely masked surface NKp30. Nevertheless and irrespective of the formal demonstration that sB7-H6 was included in serum exosomes, the impact of sB7-H6+ serum pellets on NKp30 staining and cell activation contrasted with the failure of recombinant sB7-H6 to do so. Of note, our data do not exclude that a form of sB7-H6 was not associated with exosomes. However, there are no described alternative spliced products of B7-H6, which could correspond to a soluble form lacking the transmembrane domain. Moreover, we could not detect, using our panel of mAbs, a shed form of B7-H6 which could be released from the plasma membrane by proteolytic cleavage (data not shown).
Characterization of sB7-H6 isolated from patient sera. (A) Concentrations of sB7-H6 were measured in the pellet at each round of 3 sequential centrifugations at 1200g, 12 000g, and 110 000g of B7-H6+ patient sera. Data correspond to a pool of 3 independent experiments. (B) Flow cytometric analysis of pellets purified from serum of 2 sB7-H6+ patients and 1 control individual. After purification, pellets were complexed to latex beads and stained with the indicated mAbs (black histograms); histograms obtained with isotype controls are in gray. (C) Flow cytometric analysis of NKp30 cell-surface expression on freshly isolated NK cells stained with mIgG1 isotype control (gray histograms) or NKp30 mAbs in presence of 5 μg of sB7-H6+ pellet (dashed line) or 5 μg of sB7-H6− pellet (black line). Data are representative of 3 independent experiments. (D) NKp30+ DOMsp30 reporter cells or NKp30− DO11.10 control cells (parental cells) were cocultured with K562 cells that constitutively express B7-H6, in the presence of anti-B7-H6 mAbs (17B1.3), mIgG1 isotype control, 5 μg of pellet from serum of 2 sB7-H6+ patients or 5 μg of pellet from serum of 2 sB7-H6− patients. DOMsp30 cell activation was determined by evaluating IL-2 production in the coculture supernatant in a standard CTLL-2 survival assay. Data are representative of 3 independent experiments.
Characterization of sB7-H6 isolated from patient sera. (A) Concentrations of sB7-H6 were measured in the pellet at each round of 3 sequential centrifugations at 1200g, 12 000g, and 110 000g of B7-H6+ patient sera. Data correspond to a pool of 3 independent experiments. (B) Flow cytometric analysis of pellets purified from serum of 2 sB7-H6+ patients and 1 control individual. After purification, pellets were complexed to latex beads and stained with the indicated mAbs (black histograms); histograms obtained with isotype controls are in gray. (C) Flow cytometric analysis of NKp30 cell-surface expression on freshly isolated NK cells stained with mIgG1 isotype control (gray histograms) or NKp30 mAbs in presence of 5 μg of sB7-H6+ pellet (dashed line) or 5 μg of sB7-H6− pellet (black line). Data are representative of 3 independent experiments. (D) NKp30+ DOMsp30 reporter cells or NKp30− DO11.10 control cells (parental cells) were cocultured with K562 cells that constitutively express B7-H6, in the presence of anti-B7-H6 mAbs (17B1.3), mIgG1 isotype control, 5 μg of pellet from serum of 2 sB7-H6+ patients or 5 μg of pellet from serum of 2 sB7-H6− patients. DOMsp30 cell activation was determined by evaluating IL-2 production in the coculture supernatant in a standard CTLL-2 survival assay. Data are representative of 3 independent experiments.
Discussion
B7-H6 is the most recently described member of the B7 family of cell-surface immunoreceptors.12,13,17-21 Thus far, the expression of B7-H6 has been shown to be restricted to tumor cells and absent from normal hematopoietic cells from healthy individuals at steady state.12,17 It is well known that B7 family members are induced on myeloid cells upon stimulation with infectious and proinflammatory stimuli.22-25 However, no data have been reported on the mechanisms leading to the induction of B7-H6. We have shown here that B7-H6 transcripts, B7-H6 cell-surface expression, and sB7-H6 can be induced in inflammatory conditions in vitro and in vivo. Of note, B7-H6 could not be detected at the surface of monocyte-derived DCs (moDCs).12 Functional studies demonstrated that NKp30 is crucial for the interaction of NK cells with moDCs,26 while it seems dispensable in the cross-talk with macrophages.27 Thus, 2 cell types both derived from monocytes would either express a ligand different from B7-H6 (moDCs) or, perhaps, lack NKp30 ligand expression (macrophages). Irrespective of this issue, our data indicate that B7-H6 expression is not limited to tumor cells, and reveal that nontransformed cells could be recognized as targets of NK cells through B7-H6/NKp30 interactions. They further support the concept introduced earlier for the induction of NKG2D ligands as which the ligands for NK cell–activating receptors are silenced on normal cells and induced in various conditions of cellular stress, such as infection, inflammation, and cancer. There are similarities and differences between the conditions that lead to NKG2D ligands and B7-H6 expression. On the differences, we could not detect B7-H6 induction on the surface of cells treated with DNA-damaging agents or proteasomes inhibitors that have been shown to induced cell-surface NKG2D ligands.28,29 On the similarities, TLR signaling has been shown to upregulate transcription and expression of NKG2D ligands, such as retinoic acid early inducible-1 (RAE-1) family members in mouse macrophages30 and MICA on human macrophages.31,32 These data have led us to propose some molecular basis for the cross-talks between NK cells and monocytes/macrophages during innate immune response to infections. Interactions between NK and monocytes/macrophages have been indeed documented in several inflammatory and infections conditions.33-35 As NKp30 is expressed by NK cells and B7-H6 only binds to NKp30, our present data on the induction of B7-H6 on inflammatory monocytes and neutrophils support this idea. The dissection of the mechanisms involved in these conditions is particularly relevant to sepsis, the control of which represents an important unmet medical need.
Sepsis is a clinical syndrome that complicates severe infection, and is one of the leading causes of admission to ICU with mortality rates ranging from 30% to >50%. The host response of these patients include a first phase of SIRS, characterized by an exacerbated inflammatory response, rapidly followed by a profound alteration of immunity, referred as to compensatory anti-inflammatory response syndrome (CARS). This acquired immunoparalysis is thought to render the patients more susceptible to nosocomial infections, and to lead to increased morbidity and mortality. NK cells are a major source of interferon-γ (IFN-γ), a potent inflammatory cytokine, and early depletion of NK cells improved survival in sepsis models in the mouse.36,37 We also recently observed that both NK-cell cytotoxicity and IFN-γ production were decreased in sepsis,38 consistent with similar results obtained in mice.39 Considering the sequence of opposite events that are at work during sepsis, NK cells might have a dual role in sepsis, first contributing to the amplification of the inflammatory response during the early steps of SIRS and then, impaired during CARS and thus participating to its detrimental consequences.38,40,41
Our data on B7-H6 expression during inflammation provide a novel perspective on the link between microbial infection and NK-cell activation. First, B7-H6 was selectively induced on the surface of CD14+CD16+ proinflammatory monocytes14 and neutrophils, 2 types of cells which play a central role in the onset and amplification of inflammation. In particular, CD14+CD16+ monocytes are described as the main producers of inflammatory cytokines such as TNFα and IL-1β in response to LPS.42 Several studies have reported that CD14+CD16+ monocytes are found in larger numbers in the blood of patients with acute inflammation and infectious diseases such as sepsis, tuberculosis, and rheumatoid arthritis.43-45 Second, B7-H6 could be produced as a soluble form in vitro by activated monocytes and neutrophils, and in vivo in a group of sepsis patients. Soluble forms of B7 family members as CD80, CD86, B7-H1, B7-H2, B7-H3, and B7-H4 have been detected in the serum of patients with cancer and inflammatory conditions.46-51 Although in several instances soluble B7 receptors have been shown to serve as decoy molecules to block the function of their receptors, the biological roles of these molecules remain to be dissected in detail. The present description of cell-surface and soluble B7-H6 confirms the general propensity of the members of the family to be induced in inflammatory conditions. sB7-H6 present in the serum of sepsis patients impaired the binding of anti-NKp30 mAbs to NKp30 and NKp30-dependent cell activation. Importantly, recombinant sB7-H6 was unable to impact on NKp30 expression and signaling. Thus, sB7-H6 present in patient serum is different from recombinant sB7-H6. These data support the fact that serum sB7-H6 was found in exosomal fractions and thus could be exposed as multimers with higher avidity than monomeric recombinant sB7-H6. It is, however, remarkable and still puzzling that serum sB7-H6 was restricted to patients with gram-negative infection, while no difference was observed between sepsis patients presenting gram-negative or -positive infections for mB7-H6 expression. A possible explanation could be that blood monocytes recognize gram-negative bacteria through TLR2 and TLR4 while gram-positive bacteria are recognized by TLR2. In response to ligand binding to TLR4, 2 major signaling pathways are activated. One pathway depends on myeloid differentiation marker 88 (MyD88) and is common to TLR2 and TLR4, whereas the other depends on Toll–IL-1 receptor domain-containing adaptor inducing interferon-β (TRIF) restricted to TLR4.52 Consequently, the induction of mB7-H6 could be dependent on MyD88, while the induction of sB7-H6 could be dependent on TRIF. This issue needs further investigation.
During an immune response to infection, the outcome of NK cell–myeloid cell interactions through NKp30/B7-H6 interaction could be twofold according to the membrane-bound or soluble form of B7-H6. On the one hand, 65% of patients with sepsis had circulating proinflammatory-type monocytes expressing mB7-H6, when these cells could not be detected in other SIRS patients or healthy individuals. This suggests that NK-cell activation induced by B7-H6+ monocytes could occur in these patients via NKp30 interaction. The highest mortality of patients with membrane-bound B7-H6 would suggest a negative role for NK cells in sepsis, consistent with data in the mouse.36,37 On the other hand, we also show that sB7-H6, which block mB7-H6/NKp30 functional interaction, was detected in the serum of sepsis patients. We observed a trend associating sB7-H6 with higher patient mortality (data not shown). Thus, NK-cell inhibition via sB7-H6 production would also be detrimental for the patient outcome. Based on these data, we thus propose a model in which the activation of NK cells via mB7-H6 contributes to inflammation during SIRS, but then sB7-H6 might play a negative role during CARS. This possibility would explain how 2 opposing phenomena (ie, the expression of mB7-H6 that activates NK cells and the production of sB7-H6 that impairs NK-cell activation) could both have deleterious impact on patient survival during sepsis. Our data thus prompt to perform the immunomonitoring of cell-surface B7-H6 and sB7-H6 in a large of cohort of sepsis patients over the kinetics of their clinical outcome to test whether mB7-H6 and/or sB7-H6 can help in stratifying individuals at risk of developing bad outcomes.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Louis Gastinel (Limoges University, Limoges) for help in the design of the soluble recombinant B7-H6, as well as Charlotte Viant (Centre d'Immunologie de Mareille-Luminy, Marseille) and Nicolas Manel and Clothilde Théry (Institut Curie, Paris) for help and discussions.
This work was supported by a European Research Council advanced grant (THINK), Institut National de la Santé et de la Recherche Médicale transfert (COPIO), and Agence Nationale de la Recherche and institutional grants from Institut National de la Santé et de la Recherche Médicale, Centre National de la Recherche Scientifique, and Aix-Marseille Université to the Centre d'Immunologie de Mareille-Luminy.
E.V. is a senior member of the Institut Universitaire de France.
Authorship
Contribution: J.M. and E.V. designed and analyzed the experiments and wrote the paper; J.M. performed most of the experiments; M.B. generated the anti–B7-H6 mAbs; C.C., C.P., C.F., F.V., and J.M. performed and analyzed experiments on human patients and control individuals; L.C., J.-M.F., G.T., and L.P. were responsible for the clinical studies and the statistical analysis; and D.C. and S.U. provided expression data and critical comments on the report.
Conflict-of-interest disclosure: E.V. is a cofounder and shareholder of Innate-Pharma. The remaining authors declare no competing financial interests.
Correspondence: Eric Vivier, Centre d’Immunologie de Marseille-Luminy Campus de Luminy, Case 906, 13288 Marseille Cedex 09, France; e-mail: vivier@ciml.univ-mrs.fr.