In this issue of Blood, Roberts et al report the comprehensive screening of a large cohort of Down syndrome neonates for the transient abnormal myelopoiesis (TAM) disorder based on blood cell morphology review and screening for GATA1 mutations, the signature genetic marker of TAM.1
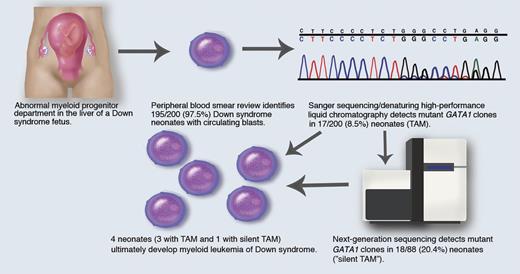
(1) Abnormal fetal myeloid progenitor department, where GATA1 mutant clones arise in the fetal liver; (2) 200 Down syndrome neonates are screened; (3) 195/200 (97.5%) have circulating blasts; (4) Sanger sequencing/denaturing high-performance liquid chromatography detects mutant GATA1 clones in 17/200 (8.5%) patients (TAM); (5) next-generation sequencing detects mutant GATA1 clones in 18/88 (20.4%) patients (“silent TAM”); (6) myeloid leukemia of Down syndrome develops in 3 TAM patients and 1 silent TAM patient. Professional illustration by Marie Dauenheimer.
(1) Abnormal fetal myeloid progenitor department, where GATA1 mutant clones arise in the fetal liver; (2) 200 Down syndrome neonates are screened; (3) 195/200 (97.5%) have circulating blasts; (4) Sanger sequencing/denaturing high-performance liquid chromatography detects mutant GATA1 clones in 17/200 (8.5%) patients (TAM); (5) next-generation sequencing detects mutant GATA1 clones in 18/88 (20.4%) patients (“silent TAM”); (6) myeloid leukemia of Down syndrome develops in 3 TAM patients and 1 silent TAM patient. Professional illustration by Marie Dauenheimer.
Children with Down syndrome (constitutional trisomy 21) not only have a high predisposition to develop acute leukemia, but also the clinical and biological features of their leukemias are very unique. In the case of myeloid leukemias (ML-DS), in which children with Down syndrome represent approximately 15% of total pediatric acute myeloid leukemia (AML) cases, these features include (1) the high prevalence of the acute megakaryocytic leukemia (AMKL) phenotype, estimated to be 500-fold higher compared with children without Down syndrome2 ; (2) detection of somatic mutations in the X-linked transcription factor gene, GATA1, almost uniformly in all Down syndrome patients with TAM and/or AMKL3 ; and (3) a prior diagnosis of TAM, which has been estimated to affect 5% to 10% of Down syndrome newborns.4
TAM (also known as the transient myeloproliferative disorder or transient leukemia), first reported in 1954, is a preleukemia disorder diagnosed in Down syndrome neonates in which peripheral blood blast cells have the morphologic, immunophenotypic, and molecular features of AMKL, with the vast majority of cases resolving spontaneously usually within 2 months.5,6 A broad spectrum of clinical findings have been reported, ranging from well-appearing, asymptomatic infants to infants with either respiratory distress and organomegaly, particularly hepatomegaly. Laboratory findings have included various degrees of leukocytosis, thrombocytopenia, and liver dysfunction with elevated bilirubin and transaminase levels. A subset of TAM patients with high-risk features (eg, hyperleukocytosis and/or hepatic failure secondary to blast cell infiltration and hepatic fibrosis) have typically been treated with low-dose cytarabine with a guarded prognosis.6 Following the clinical regression of TAM, progression to ML-DS may occur in up to 20% to 30% of patients, which requires AML-based chemotherapy treatment; ML-DS patients have an excellent overall prognosis, including individuals with a history of TAM.4,7
Based on the high predilection of patients with a history of TAM to ultimately develop ML-DS, frequent monitoring of blood counts has been recommended through the first several years of life. Detecting and defining TAM in Down syndrome neonates has remained challenging up until the current time, because it is unclear whether blood cell morphology to detect blasts is sufficient for the diagnosis as defined by the WHO classification or whether TAM should be defined by immunophenotyping of blasts for the megakaryocytic phenotype and/or detection of GATA1 mutations in blast cells.
The current study by Roberts et al is the largest and most comprehensive prospective analysis screening 200 Down syndrome neonates for evidence of TAM by reviewing complete blood counts, blood smear morphology, and analyzing for GATA1 mutations. Patients were recruited to the Oxford-Imperial Down Syndrome Cohort Study encompassing 18 United Kingdom hospitals over a 5-year period of time, which represented approximately 5% of total Down syndrome births in the United Kingdom. Two blinded observers identified that 195 (97.5%) of the patients had circulating blasts on blood smear review. This unexpected finding suggests that blood cell morphology alone is inadequate to accurately diagnose TAM in infants. GATA1 mutation analysis performed by Sanger sequencing and denaturing high-performance liquid chromatography (Ss/DHPLC) detected mutations in 17/200 (8.5%) patients who all had blasts >10%; 3 (18%) of these patients ultimately developed ML-DS.
To search further, the investigators used targeted next-generation resequencing (NGS) of GATA1 exon 2 (where the majority of mutations are identified) in 88 patients with no detectable mutations by Ss/DHPLC. NGS detected GATA1 mutations in 18/88 (20.4%) patients who had no significant difference in clinical or hematologic findings (median blasts 5% vs 4% for negative cases) compared with patients without NGS-detected mutations and would have not be diagnosed with TAM. Based on this new observation, the authors propose a new entity, “silent TAM,” to characterize patients who have no GATA1 mutations detected by Ss/DHPLC yet have mutations detected with a more sensitive screening assay with a sensitivity of ∼0.3%. One patient with silent TAM developed ML-DS, whereas leukemia did not develop in the remaining patients who had no mutant clones identified by NGS. The overall detection of GATA1 mutants in 35/200 (17.5%) patients is higher than previous estimates. Hence, second-line screening of patients by NGS, as recommended by the authors, is very important to accurately identify the at-risk population of Down syndrome neonates in whom ML-DS may develop, which would aid clinicians in providing appropriate anticipatory counseling of parents and clinical monitoring and follow-up of patients.
Five of the 35 cases had >1 GATA1 mutant clone identified, suggesting that the fetal environment and the potential role of chromosome 21–localized genes lead to a mutator phenotype and GATA1 mutations may confer a survival advantage to the mutant clones.8 Abnormalities in Down syndrome fetal hematopoiesis have also been linked to the myeloid progenitor compartment, including an increased number of megakaryocyte-erythroid progenitors, and GATA1 mutant clones have also been detected in Down syndrome fetal liver samples as early genetic “hits” in leukemogenesis.9,10
An unanswered question is whether all ML-DS cases arise from a preceding case of TAM that may not have been identified in the neonatal period. The findings of this study will serve as a model for other large population-based screening studies of Down syndrome neonates for TAM. By identifying an at-risk group of patients, closer clinical monitoring of TAM patients may also reveal whether exogenous factors or second postnatal genetic hits play a role in the progression to leukemia. A small proportion of cases, however, would likely be missed, because GATA1 mutations can also be detected in neonates with Down syndrome mosaicism who lack the phenotypic features of Down syndrome that would identify them for screening.
An ultimate study would encompass prospective screening of all Down syndrome neonates for TAM and the use of a therapeutic intervention (repetitive courses of very-low-dose cytarabine administered on an outpatient basis) to determine if this can eradicate the GATA1-containing mutant preleukemia clones and prevent the development of ML-DS altogether.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal