Abstract
The treatment success in patients (pts) with acute myeloid leukemia (AML) is very heterogeneous. Cytogenetic and molecular alterations present at diagnosis are strong prognostic factors, which have been used to individualize treatment. As shown by several groups, the subgroup of pts with deletion of the short arm of chromosome 17 are at high risk for treatment failure (e.g. Seifert, Leukemia 2009), which persists even after allogeneic hematopoietic stem cell transplantation (HSCT) (Middeke, Blood 2012; Mohr, Br J Haematol 2013). Besides allelic loss of TP53 located on the short arm of chromosome 17, other mechanisms of inactivation have been shown for this key tumor suppressor gene, most importantly missense point mutations or small deletions. These alterations have also been linked to poor outcome in AML after chemotherapy (Grossmann, Blood 2012). Here, we studied the impact of TP53 mutations on the outcome of AML pts with adverse cytogenetic risk treated with HSCT.
We selected AML pts with complex karyotype (CK), monosomy 7, monosomy 5/del5q and/or abnl(17p) who had received HSCT within 3 randomized controlled trials (NCT numbers 00180115, 00180102, and 00180167). All pts were treated with intensive induction chemotherapy and HSCT according to a risk adapted strategy.
Complete sequencing of the TP53coding region was done using next generation sequencing (NGS) on a 454 GS Junior instrument (Roche) using the IRONII-study amplicon panel. Amplicons were generated from genomic DNA isolated at the time of diagnosis. Data analysis was done using the Sequence Pilot software package (JSI Medical Systems), a 10% cut-off was used for mutation calling.
Nonsynonymous mutations were classified into bi-allelic TP53 mutations if detected allelic frequency as determined by NGS was >50% and mono-allelic TP53 mutations for frequencies between 10% and 50%. All samples with synonymous mutations or no detectable mutations according to the predefined cut-off of 10% were classified as TP53wild type (wt).
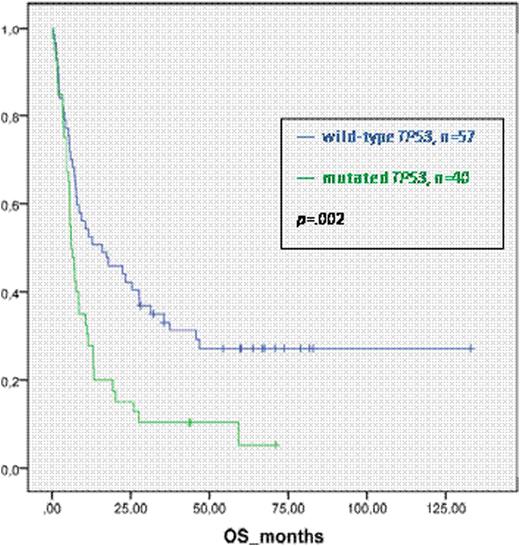
Overall survival (OS), event-free survival (EFS), cumulative incidence of relapse (CIR) and non-relapse-mortality (NRM) after HSCT were analyzed according to the mutational status.
Samples from 97 pts with AML were analysed, the median age was 51 years (range 18 to 67), 83% suffered from de novo AML, while 13% had sAML and 3% therapy-related myeloid neoplasms. CK and monosomal karyotype (MK) were present in 61% and 42% of the pts, respectively. Twenty-nine pts (30%) had abnl(17p) detected by conventional karyotyping or FISH analysis. Twenty-six pts (27%) were treated with standard myeloablative conditioning (MAC) regimens while the remaining pts received reduced intensity conditioning (RIC). Donors were siblings in 36 pts (37%) and matched or mismatched unrelated donors in all other pts.
Overall, TP53 mutations were found in 40 pts (41%). Twenty-eight (29%) pts had a bi-allelic TP53 mutation while 12 (12%) pts had a mono-allelic TP53 mutation. We identified 15 pts with TP53 mutations without abnl(17p). Four pts with abnl(17p) had wt TP53. Pts with TP53 mutations were significantly older than pts with wt TP53 AML (median age 55 vs. 43, p=.004). Donor type, type of conditioning and the rate of transplantation in first complete remission were not statistically different among pts with or without TP53mutations.
With a median follow up of 67 months the three-year probabilities of OS and EFS for pts with wt TP53 were 33% (95% CI, 21% to 45%) and 24% (95% CI, 13% to 35%) compared to 10% (95% CI, 0% to 19%) and 8% (95% CI, 0% to 16%) (p=.002 and p=.007) for those with mutated TP53, respectively. CIR at three years was 42% for pts with wt TP53 and 60% for those with mutated TP53 (p=.05). NRM was not different in both groups. In multivariate analysis including age, donor type (sibling vs. all other), type of conditioning (RIC vs. MAC) and disease status (CR1 vs. advanced disease) only the TP53-mutation status had a significant influence on EFS (HR=1.72; p=.03). In our analysis, classification according to MK did not significantly influence OS, EFS, CIR or NRM.
In this cohort of pts with cytogenetic adverse risk abnormalities, who had received HSCT, TP53 mutations were present in 41% of the pts. OS and EFS were significantly worse in pts with mutated TP53. Mutational analysis of TP53 might be an important additional tool to predict outcome after HSCT in pts with adverse karyotype AML.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal