Abstract
Detection of minimal residual disease (MRD) by t(14;18) Real-Time Quantitative (RQ) PCR is an important tool during clinical follow-up in patients (pts) with follicular lymphoma (FL). However, only the major breakpoint region (MBR) and minor cluster region (mcr) breakpoints have been currently exploited as MRD targets. Therefore, only 50% to 65% of pts can be assessed by t(14;18) RQ-PCR. Alternative targets such as the immunoglobulin heavy chain variable region (IGH) can be used only with limitations due to somatic hypermutation. IGH-based next-generation sequencing (NGS) might provide an alternative approach and provide increased sensitivity, specificity, accuracy and reproducibility. We performed a comparison of the two approaches in 29 FL pts.
Overall, 206 samples (85 bone marrow, 114 peripheral blood, 6 stem cell aliquots and one lymph node sample) were investigated from 29 FL pts in which RQ-PCR based MRD analysis had been performed in the context of prospective clinical trials. Overall, 33 dx and 173 follow-up (FU) samples were analyzed. 23/29 pts had a PCR detectable t(14;18) rearrangement, 5 pts had a clonal IGH rearrangement only and one patient had no marker by consensus PCR. NGS was performed at Sequenta in South San Francisco. Using universal primer sets, we amplified IGH variable, diversity, and joining gene segments from genomic DNA. Amplified products were sequenced to obtain a high degree of coverage and analyzed using standardized algorithms for clonotype determination. Tumor-specific clonotypes were identified for each patient based on their high-frequency in the dx sample and then quantitated in FU samples.
Overall comparability of MRD results by RQ-PCR and NGS was assessed using correlation analysis. A positive/negative discordance between two results was defined as major when the positive result was >1E-05 and minor when ≤1E-05; a quantitative discordance was defined as the presence of two positive results with a quantitative discrepancy >1 log.
All 29 pts were evaluable with at least one method. IGH NGS identified an index clone in 22/29 cases. Seven patients pts were only quantifiable by t(14;18) RQ-PCR while IGH NGS did not identify an index clone for sequencing. In all but one dx sample demonstrated low level lymphoma infiltration with MRD level below 10-3. In one of these cases, IG kappa could be successfully sequenced for MRD indicating that not only low level MRD but also somatic mutation of IGH is a potential pitfall for MRD detection by NGS in FL.
15 pts were evaluable for MRD by t(14;18) RQ-PCR and NGS. In 97 FU samples, a significant concordance between MRD methods could be demonstrated (r2=0.80) (p<0.0001). Of these samples, 44 were MRD positive and 45 were MRD negative with both tools. A quantitative discordance occurred in 12/44 MRD positive samples, where MRD was higher in 7 samples and lower in 5 samples by NGS. A major discordance occurred in 4 samples where t(14;18) RQ-PCR was positive and NGS was negative. A minor discordance was detectable in 4 samples where in 2 samples t(14;18) RQ-PCR was positive and NGS was negative, and in the other 2 samples the opposite was correct.
In 5 t(14;18) negative cases and one with an unusual large t(14;18) rearrangement MRD was quantified by IGH-RQ-PCR using cloned plasmids and IGH NGS. In all cases both methods also showed excellent concordance.
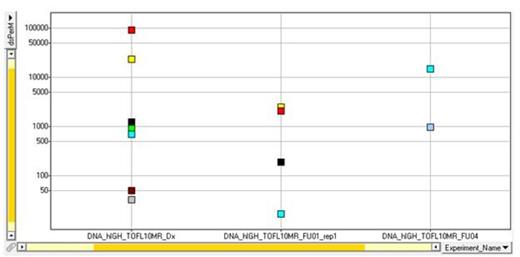
One advantage of the IGH NGS approach is its ability to detect different IGH sequences generated by the somatic hypermutation process. We have seen in some patients a dynamic picture of the IGH clonotypes with the rise and fall of different clonotypes at different time points (Fig 1).
New IGH clonotypes generated by somatic hypermutation appear over time. Each color is a different clonotype, and shape reflects number of single base mutations from the index clone.
New IGH clonotypes generated by somatic hypermutation appear over time. Each color is a different clonotype, and shape reflects number of single base mutations from the index clone.
NGS represents a feasible tool for IGH-based MRD monitoring that allows analysis of a larger group of FL pts. Our results show that the two methods have a high level of correlation. Lymphoma infiltration of dx samples and somatic mutation of IGH is a critical point for identification of the tumor-specific clonotypes by NGS, therefore different MRD methods should complement each other to allow MRD assessment for the majority of pts. Furthermore IGH NGS sequencing has the potential to detect and track IGH evolution in FL.
Kneba:Roche: Consultancy, Research Funding. Faham:Sequenta, Inc.: Employment, Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal