Abstract
We performed gene expression profiling of 36 symptomatic and asymptomatic WM patients, 13 IgM-MGUS cases and 7 healthy subjects as controls (CTRL) using Affymetrix GeneChip Human Genome U133 Plus 2.0 Array.
Bone marrow (BM) CD19+ cells (n=36) and BM CD138+ (n=32) cells were isolated from WM, BM CD19+ cells (n=10) and BM CD138+ (n=10) cells were selected from IgM-MGUS while BM CD19+ cells (n=7) and BM CD138+ (n=7) cells were isolated from CTRL.
Data were preprocessed and normalized using RMA and ComBat. Selection and data analysis were performed using ANOVA followed by t-test, both adapted for microarrays and functional pattern discovery (Di Camillo et al., PLOS ONE 2012).
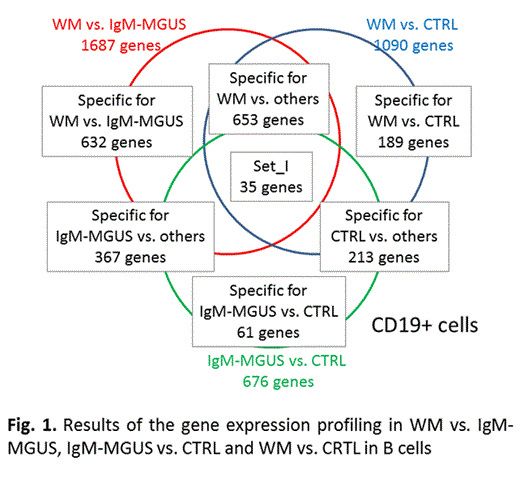
The comparison between WM, IgM-MGUS and CRTLs CD19+ cells highlighted many differentially expressed genes. The comparison across all CD19+ cells identified 35 genes selected in all the comparisons (SET I); in particular, ADARB1 (alternative splicing), FANCI and ABCB1 (cell cycle), CRY1 and SATB1 (negative regulation of transcription) and ADAM23 (cell adhesion) were progressively underexpressed in CTRL vs. IgM-MGUS vs. WM.
The comparison between WM vs. IgM-MGUS CD19+ cells demonstrated that a large number of genes (n=1687) were differently expressed, of which 632 specific for the WM vs. IgM-MGUS comparison and enriched for immune response, signal transduction, MAPK pathway, angiogenesis, cell cycle, transcription, alternative splicing and protein phosphorylation annotations. Among these, CXCL4, CXCL4L1, CD86 regulating immune response were underexpressed in WM B cells with the fold changes of 5.41, 4.11 and 2.93, respectively. RGS18, P2RY12, RAB27B (signal transduction) were downregulated in WM B cells with 5.42, 3.64, 3.10 fold changes, respectively.
Regarding MAPK pathway, SYK was overexpressed while ADAM29, EGF, THBS1, PPM1L, FGFR1 were underexpressed in WM B cells. Genes involved in angiogenesis such as MEIS1, ANGPT1 were underexpressed, whereas PRKD2, CASP8, KRIT1 were overexpressed in WM B cells. SEPT8, SEPT6 were upregulated while PRKAR2B, SEPT5, PRR5 were underexpressed in WM B cells in the cell cycle. 104 genes regulating transcription were differently expressed in WM B cells and among them, 38 were zinc finger proteins. Among genes involved in protein phosphorilation, ITGB3, P2RY1, CTDSPL, KALRN, DAB2 were underexpressed while SGK494 and SIK3 were upregulated.
The comparison between WM vs. CTRL as well as that between IgM-MGUS vs. CTRL of B cells showed that cell cycle, transcription, and cell adhesion differentiated WM and IgMMGUS in respect to CTRL. Of note, genes belonging to the cell cycle: ASPM, CENPE, ESCO2, CDCA2, MKI6, BUB1, CHEK1, CASC5 were progressively underexpressed in CTRL vs. IgMMGUS vs. WM with fold changes ranging between 3 and 5.
LEF1, SSBP2, MYB, CDCA7L genes regulating transcription were progressively underexpressed in CTRL vs. IgMMGUS vs. WM with fold changes ranging between 2.5 and 4. RAG1 and IL4R (immune response) were underexpressed with high fold changes of 6.18 and 3.99 in WM vs. CTRL and with fold changes of 2.89 and 2.18 in the comparison of IgMMGUS vs. CTRL B cells, respectively.
A comparison across all CD138+ cells (Set I) showed that ESRP1 (RNA splicing), IGHD (immune response), TJP1 (apoptosis), and CADPS2 (protein transport) were progressively underexpressed in CTRL vs. IgMMGUS vs. WM with high fold changes ranging between 4 and 7.
The comparisons between WM vs. IgM-MGUS and WM vs. CTRL highlighted that GBA3 belonging to sphingolipid pathway, SEPT10 and PARD3 (cell cycle), IGLC1 (immune response), NRG3 (regulation of cell growth) were underexpressed in WM. No genes were specific for the IgM-MGUS vs. CTRL comparison.
In conclusion, the study demonstrated that many genes involved in alternative splicing, regulation of transcription, protein phosphorilation, cell cycle, and immune response were progressively underexpressed in CTRL vs. IgM-MGUS vs. WM in B cells.
Conversely, the comparison between WM vs. IgM-MGUS as well as WM vs. CTRL plasma cells demonstrated that 12 genes were underexpressed in WM. In addition, microarray results showed that IgM-MGUS plasma cells seem to be similar to the normal cell counterpart.

No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal