Abstract
The assessment of minimal residual disease (MRD) is a key component of prognosis and monitoring in acute lymphoblastic leukemia (ALL) and mantle cell lymphoma (MCL). Allele-specific oligonucleotide (ASO)-PCR can be used to assess MRD; however, this technique requires preparation of clonotype-specific primers for each patient, which is laborious and time-consuming. We demonstrated the utility of sequencing-based MRD assessment in ALL (Faham et al., Blood 2012). This quantitative approach relies on amplification and sequencing of immunoglobulin and T-cell receptor gene segments using consensus primers and can address some of the limitations associated with traditional MRD detection. Here we compared the ability of the sequencing and ASO-PCR methods to identify clonal cancer gene rearrangements at diagnosis and evaluated the concordance of MRD detection in bone marrow (BM) or blood samples from 37 patients (pts) with ALL and 22 pts with MCL entered onto prospective CALGB treatment trials, 10403 and 59909 (Alliance), respectively .
Using the quantitative ASO-PCR and sequencing assays, we analyzed diagnostic blood and BM samples from ALL pts for clonal rearrangements of immunoglobulin (IGH-VDJ, IGH-DJ, IGK) and T cell receptor (TRB, TRD, TRG) genes. We then assessed MRD at the IGH and/or TRG locus in 84 follow-up samples from ALL pts. Similarly, we analyzed samples from 22 MCL pts for immunoglobulin (IGH-VDJ, IGH-DJ, IGK) clonal gene rearrangements and measured MRD at the IGH and/or IGK locus in 114 follow-up samples. Sensitivity of the ASO-PCR vs sequencing was 1 X 10 4-5 vs 1 X 106, respectively. Concordance between sequencing and ASO-PCR MRD assessment on serial samples collected during and post-treatment was evaluated across all pts and within disease groups using concordance correlation coefficients for repeated measures (CCC-RM). Concordance in identification of detectable vs. undetectable MRD by both methods was also evaluated using Kappa statistics.
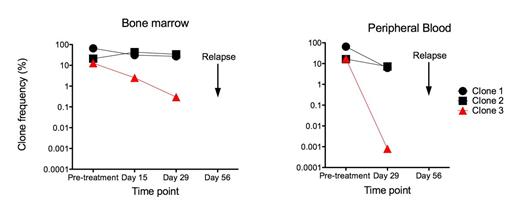
Using the sequencing platform, high frequency clonal rearrangements were observed in at least two receptors in 97% and 95% of pts with ALL and MCL, respectively. Selected ALL samples were known to have IGH-VDJ or TRG clonal rearrangements by ASO-PCR; however, sequencing revealed additional clonal rearrangements in 36/37 (97%) pts with ALL. Good concordance was observed with identification of MRD positive vs. negative between the methods (K=0.62; p<0.0001), where better agreement was observed within the ALL (K=0.63) than within the MCL (K=0.59) cohorts. Across both diseases, the sequencing method proved a more sensitive measure and CCC-RM was 0.82, reflecting the fact that 28 pts with undetectable MRD by ASO-PCR methods had low detectable MRD measures by the sequencing approach. The sequencing platform also provided new insights into kinetics of relapse. In one ALL case (Fig 1), we observed three high frequency IGH-VDJ clones, two of which are related based on VDJ replacement model. Dramatic reduction in frequency of one clone (clone 3, Fig 1) was observed following initial induction chemotherapy in both BM and blood. Only this single clone was monitored using ASO-PCR, and the patient appeared to be in molecular remission. However, the other two high frequency clones were chemo-resistant, and sequencing MRD monitoring revealed no reduction in clone frequency in either the BM or blood. The patient experienced early relapse at day 56, which may have resulted from the expansion of a clone carrying one or both of the clonal sequences that was not monitored by ASO-PCR. Thus, the sequencing method can be used to monitor response to treatment at the individual clone level.
This study demonstrates concordance between identification of detectable MRD in ALL and MCL by sequencing and ASO-PCR Methods. The sequencing approach offers improvements over ASO-PCR including the ability to monitor multiple clonal sequences, faster turnaround time (results in 1 week), and greater sensitivity. The clinical significance of this greater sensitivity remains to be tested prospectively and must be correlated with clinical results. Nevertheless, the sequencing method represents an alternative approach for clinical MRD monitoring which could fundamentally improve the ability to monitor disease progression and recurrence in patients with lymphoid malignancies.
Carlton:Sequenta, Inc.: Employment, Equity Ownership. Weng:Sequenta, Inc.: Employment, Equity Ownership. Faham:Sequenta, Inc.: Employment, Equity Ownership, Membership on an entity’s Board of Directors or advisory committees.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal