Abstract
Diamond-Blackfan anemia (DBA) is a bone marrow failure syndrome associated with ribosomal protein (RP) deficiency caused by mutations in RP genes. 25% of patients carry mutations in the RPS19 gene. A salient feature of DBA is proliferative arrest of erythroid progenitors. It is still not clear how defects in RPs, which are essential to all cell types, affect the erythroid compartment more severely than the others. Today, DBA patients are treated with glucocorticoids and/or blood transfusions, often for long periods, causing severe adverse effects. We hypothesize that novel drugs with more disease-specific therapeutic mechanisms can be developed and that such drugs will be superior to current therapies. Towards this aim we have developed a method for screening chemical libraries to identify novel drug candidates.
Use of DBA patient cells is not feasible as their limited availability severely limits the size of the applied screening library. Immortalized cell lines are also less suitable since the disease mechanism involves TP53 activation and most cell lines have perturbations in the TP53 pathway. Hence, primary c-Kit+ E14.5-15.5 fetal liver erythroid progenitor cells from a mouse model of DBA with doxycycline inducible expression of rps19-shRNA were used (Jaako et. al. Blood, 2011). The DBA phenotype is induced by adding doxycycline to the culture medium which reduces erythroid proliferation >80%. Rescue of this proliferation defect is a simple and relevant readout for large-scale screening.
Assay development in 96 well microtiter plates allowed rational liquid handling. Firstly, the proliferation readout method was optimized. Use of CellTiter-Glö Luminescent Cell Viability Assay generated data with superior linear correlation with cell numbers compared to Prestobluë and high content screening microscopy approaches. To enable large-scale screening, the assay was further optimized for cell numbers per well, media composition, culture duration, doxycycline concentration and timing of induction and of addition of test chemicals. We also controlled for evaporation during incubation to significantly reduce plate edge effects. During these optimizations, luminescence readout from uninduced cells was set to represent 100% rescue and readout from doxycycline-induced controls to represent 0% recue. This allows reliable normalization between plates and gives toxic chemicals a negative value and chemicals rescuing proliferation a positive value with 100% meaning complete rescue. To objectively quantify how changes of different parameters improved variability in both induced and uninduced cells, we calculated the Z factor as described by Zhang et. al. (JBS, 1999). The Z factor takes into consideration both the range and variability of data to calculate the suitability of an assay for high throughput screening. It is represented as:
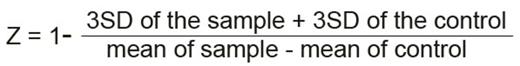
(SD: Standard Deviation). Where Z value is meaningful in the range of -1 < Z ² 1. The larger the value of Z, the higher is the data quality and a Z-factor above 0.5 is considered very robust.
After assay development we arrived at the conditions shown in the table, resulting in the Z factor of 0.7:
| Culture medium | 100 ng/ml mSCF, 2 U/ml Epo, 100 nM dexamethasone in serum-free medium. (doxycycline: 0.5 μg/ml). |
| Culture days | 4 (addition of test chemicals 24 hours after doxycycline induction) |
| Cells per well | 2000 (murine cKit+ fetal liver cells with inducible rps19-shRNA) |
| Readout | Luciferase based ATP detection (CellTiter-Glo¨). |
| Culture medium | 100 ng/ml mSCF, 2 U/ml Epo, 100 nM dexamethasone in serum-free medium. (doxycycline: 0.5 μg/ml). |
| Culture days | 4 (addition of test chemicals 24 hours after doxycycline induction) |
| Cells per well | 2000 (murine cKit+ fetal liver cells with inducible rps19-shRNA) |
| Readout | Luciferase based ATP detection (CellTiter-Glo¨). |
For screening involving small molecule based libraries we added the compounds 24 hours after cell seeding and doxycycline addition, which allows for the induction of the proliferation perturbed phenotype. We have pharmacologically validated the assay by testing and quantifying the impact of IL-3, a cytokine known to have a positive effect on erythroid progenitors. IL-3 had a 25% rescue value in our assay. Ongoing experiments (>11,000 compounds screened to date) show that more than 50% of compounds with >20% rescue score in the screen could be confirmed to rescue proliferation in dose-dependent experiments.
In conclusion, we have established a robust scalable assay for screening molecules that rescue proliferation arrest caused by Rps19-deficiency in mouse erythroid progenitors. We are currently using this assay to screen small molecule libraries in our search for novel tool compounds for DBA research and drug candidates for DBA treatment.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal