Abstract
Memory B cells (MBCs) remain viable in a non-proliferative state for years. These cells express genes involved in cell survival and anti-apoptotic factors, while repress the expression of cell cycle regulatory genes. During their differentiation into plasma cells (PCs), these cells develop an opposite gene expression pattern, with a higher expression of genes implicated in cell proliferation and activation, and a lower expression of survival genes. In multiple myeloma (MM) the PCs accumulate into the bone marrow due, at least in part, to failure of pro-apoptotic mechanisms normally expressed in PCs. In the present study we analyzed the gene expression patterns of MBCs and PCs from healthy donors and patients with MM, in order to determine whether or not myelomatous PCs share characteristics of MBCs and/or normal PCs, and to identify possible genes related to the pathophysiology of the disease.
MBCs were obtained by immunomagnetic separation from buffy coats of 5 aged healthy donors. Likewise, PCs were isolated from bone marrow of 6 healthy donors and of 5 patients with MM. Using microarray techniques we analyzed the expression of 45000 genes in all samples. We performed unsupervised hierarchical cluster of gene expression data using the average linkage and the Euclidean distance. To identify differentially expressed genes among experimental groups we applied non-parametric Kruskal Wallis test. The differences in expression with a p value < 0.05 were considered significant. All analyses were performed using the Multi-experiment Viewer 4.7.1 software.
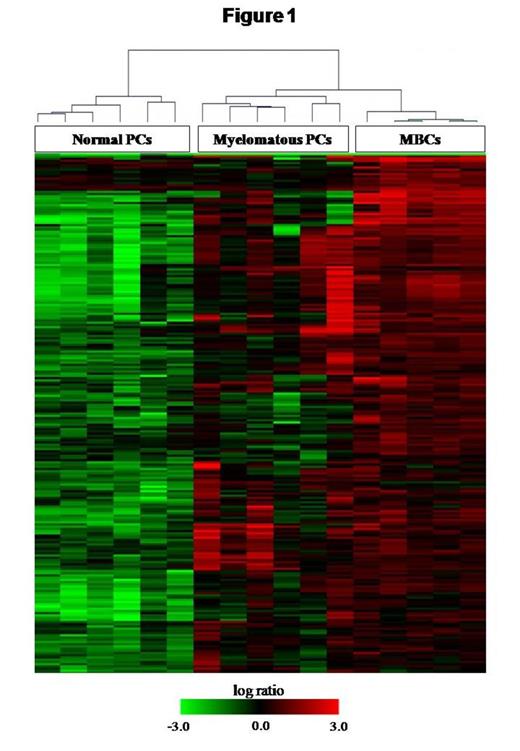
From the hierarchical cluster obtained we clearly identified two groups, one which included normal PCs samples and the other one containing myelomatous PCs and MBCs. Interestingly, myelomatous PCs displayed intermediate features between MBCs and normal PCs (Figure 1). We found 5159 genes differentially expressed between normal and myelomatous PCs. Among these, we identified 3455 genes which displayed a similar expression pattern between MBCs and myelomatous PCs, including caspases inhibitors, MAP kinases, ubiquitins, transcription and translation factors.
Myelomatous PCs display an intermediate gene expression pattern between normal PCs and MBCs. These cells display high expression of genes involved in cell survival that should be normally inactivated in the transit of MBC to a normal PC, so that its expression pattern is closer to a MBC than a normal PC.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal