Abstract
The monoclonal antibodies against CD20 antigen (rituximab and ofatumumab) have been developed and used in a clinic as a therapeutic strategy in B-cell malignancies. These antibodies eliminate B cells by triggering indirect effector mechanisms of the immune system, namely complement-dependent cytotoxicity (CDC), antibody-dependent cellular cytotoxicity (ADCC), or immunophagocytosis. Unfortunately, in some patients the resistance to anti-CD20 therapy has been detected. Therefore the molecular mechanisms responsible for the therapy failure should be urgently elucidated. One of the major mechanisms responsible for the resistance to anti-CD20 therapy seems to be the reduced level of CD20 antigen on the surface of tumor B-cells. We have previously discovered that CD20 expression is strictly dependent on the activity of Src family tyrosine kinases and AKT kinase. We have noticed that treatment of B cells with Src family inhibitors or AKT inhibitors leads to a dose-dependent reduction of CD20 at both transcript and protein level. On the other hand the overexpression of constitutively active AKT (CA-AKT) leads to a significant increase in CD20 mRNA level. To uncover the transcriptional mechanisms governing the CD20 expression we employed the construct encoding the promoter region of CD20 cloned upstream of the firefly luciferase gene. The truncated and mutated versions of the CD20 promoter were used in the luciferase assays to elucidate the role of particular transcription factors binding sites in the regulation of CD20 expression.
The aim of this study was to explore the molecular mechanisms governing the transcriptional regulation of CD20 expression in lymphoma cells as a potential explanation of the resistance to anti-CD20 therapy.
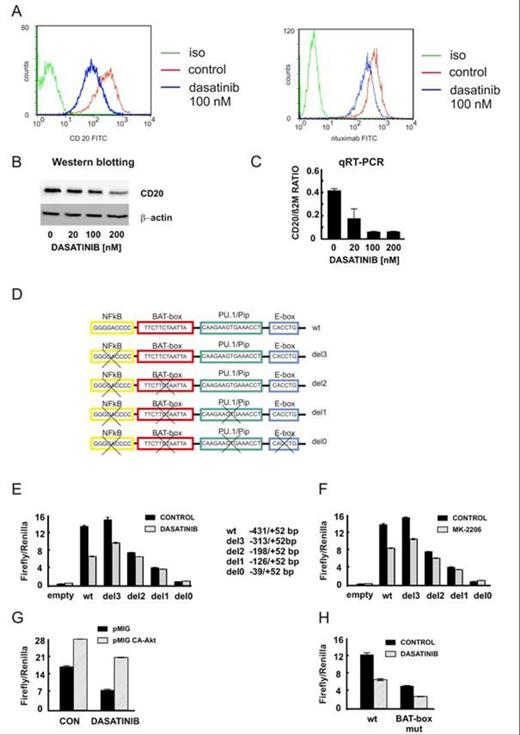
In the initial experiments we observed a significant reduction of CD20 protein level in Raji cells treated with Src family tyrosine kinase inhibitor, dasatinib (Fig. 1A,B). This reduction correlated with the impaired binding of anti-CD20 monoclonal antibodies as estimated by FACS analysis. Quantitative PCR analysis revealed that the transcriptional regulation is the major mechanism responsible for the reduction of CD20 level upon dasatinib treatment (Fig. 1C). Consistently, the exogenously expressed CD20 under the control of CMV promoter was not sensitive to dasatinib treatment. To further elucidate the mechanism of transcriptional regulation of CD20 we performed the luciferase assays to estimate the activity of CD20 promoter and its truncated forms (Fig. 1D). Dasatinib or AKT kinase inhibitor (MK-2206) strongly decreased the activity of CD20 promoter (Fig. 1E,F) while the overexpression of CA-AKT partially blocked the inhibition caused by dasatinib (Fig. 1G). Using the truncated versions of the CD20 promoter we found that lack of the region (-313/-198) made the promoter insensitive to dasatinib treatment. Since this particular region is known to contain a putative Octamer transcription factor binding site (BAT-box, Thevenin et al., 1993), we introduced mutations in the BAT-box sequence. Although basal promoter activity was indeed decreased (Fig. 1H), dasatinib was equally effective in reducing the activity of both wild-type and mutated CD20 promoter. Collectively, our results indicate that Octamer transcription factor is an important regulator of basal CD20 expression, but it is not the major mediator of the effects caused by Src family inhibitors.
Our studies indicate that the Src family tyrosine kinases and AKT kinase are involved in the transcriptional regulation of CD20 antigen in lymphoma cells. The activity of CD20 promoter is significantly reduced upon treatment with Src family inhibitors, namely dasatinib. The particular region of CD20 promoter (-313/-198) was identified as the major region sensitive to dasatinib treatment. The transcriptional machinery responsible for the reduction of CD20 expression by dasatinib needs further investigation since the expected Octamer transcription factor does not mediate the effects caused by dasatinib.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal