Abstract
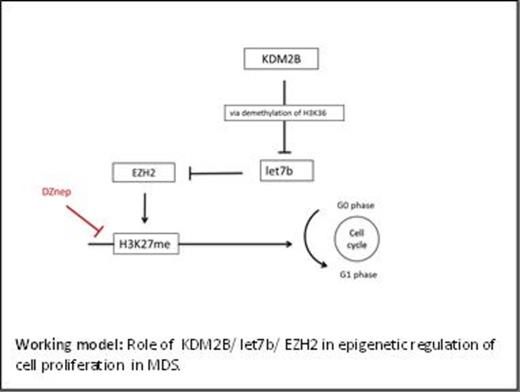
DNA and histone methylation as well as microRNA (miR) expression are dysregulated in MDS. The histone demethylase KDM2B suppresses let7b, which post-transcriptionally controls EZH2, a histone-lysine N-methyl transferase that methylates lysine 27 on histone 3. We hypothesized that epigenetic dysregulation in MDS involves the KDM2B/ let7b/EZH2 axis (Figure). DZNep (cyclopentanyl analog of 3-deazaadenosine) is a novel compound, which results in loss of lysine27 methylation, re-expression of epigenetically silenced genes, and decreased cell proliferation.We propose that DZNep will bypass cellular dysregulation via the KDM2B/let7b/EZH2 axis and thereby enhance therapeutic efficacy when combined with azanucleosides.
CD34+ MDS marrow cells were obtained by density separation and enriched by magnetic-activated cell sorting (MACS; Milteny Biotec, Auburn, CA); purity 95% to 98% by flow cytometry. For NanoString nCounter miR array total cell derived RNA or synthetic miR pools (IDT; 30 pmol per oligonucleotide) were used as input. Nano-string data were validated by real-time polymerase chain reaction (PCR). RNA was extracted using the RNeasy Mini kit (QIAGEN, Valencia, CA). Primers for let7b, U6, KDM2b and EZH2 were synthesized by Applied Biosystems. Total RNA was reverse transcribed with Taqman MiR Reverse Transcriptase kit (Applied Biosystems, Carlsbad, CA). Potential functional roles of KDM2B, let7b and EZH2 were determined in myeloid cell lines MDS-L, PL-21, KG1a,OCI-AML3 and ML1. Conditional knock-down of KDM2B was achieved with 2 short hairpins (open-biosystems). Expression of KDM2B was forced using pCMV-Myc-KDM2B (gift of Dr. Tzatsos) in cell lines with low/absent constitutive levels of KDM2B and high levels of let7b. miRZip plasmids (miRZip anti-sense) were used to knock-down let7b; pre-BAB-let7b (origene) served to overexpress let7b. Gene expression, protein levels and cell proliferation were analyzed by RQ-PCR, western blot and BrdU uptake. To determine the impact of modified let7b and KDM2B expression on treatment responses, MDS-L cells (high constitutive levels of EZH2) were treated with DZNep and methyltransferase inhibitor, 5-azacytidine, and subjected to the same analyses.
Let7b levels were significantly higher in primary CD34+ MDS marrow cells (n=44) than in healthy controls (p= 0.0304), while KDM2B and EZH2 expression was reduced (n=21, p<0.0126, p<0.0001, respectively). Overexpression of let-7b in cells with low constitutive levels (PL21, ML1 and KG1a) reduced EZH2 and KDM2B protein levels, and decreased S-phase while increasing G0/G1 cells (p=0.0005); this was associated with a decrease of H3K27me3 and cyclin D1. Silencing of KDM2B in cells with high (MDS-L) and low (KG1a) constitutive levels of let-7b resulted in increased levels of let7b. This increase in let-7b was associated with reduced levels of EZH2 protein and H3K27me3. These findings were accompanied by reduced proliferation (fewer cells in S-phase, more cells in G0/G1). Addition of DZNep to the myeloid cell lines under study resulted in a decrease of let-7b expression, increased levels of EZH2, but reduced cell proliferation. When DZNep (1 uM/ml) was combined with 5-azacytidine (AZA, 5 uM/ml), levels of EZH2 increased in parallel with reduction of H3K27me3 and cell proliferation. KG1a cells overexpressing let-7b (to mimic conditions in primary MDS cells) showed further increase of EZH2 in response to DZNep and AZA.
The KDM2B/ let7b/EZH2 axis is involved in epigenetic regulation in MDS. KDM2B, via let-7b/EZH2, promotes transcriptional repression in myeloid cell lines and primary MDS cells; KDM2B controls epigenetic modifications and cell proliferation. DZNep bypasses the “inhibitory” KDM2B/let7b/EZH2 axis by preventing H3K27 methylation and reducing cell proliferation, independent of EZH2 expression. Thus, the use of DZNep would add histone demethylation to the effect of azanucleosides on DNA methylation, thereby enhancing the therapeutic success in MDS.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal