Abstract
Mutations in the BCR-ABL1 tyrosine kinase domain (TKD) are regarded as the most important mechanism of resistance to tyrosine kinase inhibitors (TKIs) in patients with Ph-positive leukemias. The occurrence of two or more mutations on the same DNA molecule, the so-called compound mutations, can be associated with particularly high resistance to multiple TKIs. Recent reports indicate that the frequency of compound mutations is rather high, thus rendering their reliable detection an important diagnostic challenge 1,2. Analysis of PCR amplicons of the BCR-ABL1 TKD by next generation sequencing (NGS) has become the method of choice for sensitive detection of compound mutations. This approach is, however, hampered by the requirement of 3-4 overlapping amplicons to cover the entire TKD due to the limited read length offered by most current NGS technologies. This prevents the assignment of nucleotide substitutions located on different amplicons to the same TKD/DNA molecule, and therefore requires additional laborious steps to facilitate unequivocal identification of such constellations. To overcome this limitation, we have established a long-range NGS approach on the FLX instrument (Roche) permitting the coverage of the entire TKD length of ∼0.9 kb in a single read. By testing a series of individual and consecutive specimens derived from five patients with chronic myeloid leukemia, we demonstrate that long-range NGS analysis readily permits the identification of mutations and their assignment to the same or to separate subclones at a limit of sensitivity comparable to NGS-based sequencing of shorter amplicons. In addition to the detection of individual and compound mutations, this approach also facilitates an interpretable documentation of insertions and deletions in the TKD. To address the possibility of artifacts inherent in the technique that could lead to incorrect identification of single and compound mutations, the NGS findings were reevaluated by independent technical approaches. Point mutations were confirmed by Sanger sequencing, LD-PCR 3 and pyrosequencing 4. In select cases, PCR amplicons of the BCR-ABL1 TKD derived from individual specimens were subcloned into pGEM®T easy plasmids, and >100 clones were subjected to analysis by Sanger sequencing. The observations made by NGS analysis including various single mutations (e.g. G250E, Y253H, T315A, F317I, Q252H, T315I), compound mutations (e.g. G250E/Y253H, G250E/T315A, G250E/F317I), and combinations of point mutations with small insertions or deletions (e.g. E459K/C475fs, Q252H/R362fs, T315I/R362fs) as well as large deletions involving multiple exons, could be confirmed in individual clones by Sanger sequencing, thereby documenting the reliability of the long-range NGS technology. The technical advancement presented therefore provides an economic approach to the identification of compound mutations and other genetic alterations in the entire BCR-ABL1 TKD, thus extending the diagnostic armamentarium for rapid assessment of impending resistant disease.
1. Khorashad JS, Kelley TW, Szankasi P, et al. BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: frequency and clonal relationships. Blood. 2013;121(3):489-498.
2. Soverini S, De Benedittis C, Machova Polakova K, et al. Unraveling the complexity of tyrosine kinase inhibitor-resistant populations by ultra-deep sequencing of the BCR-ABL kinase domain. Blood. 2013.
3. Preuner S, Denk D, Frommlet F, Nesslboeck M, Lion T. Quantitative monitoring of cell clones carrying point mutations in the BCR-ABL tyrosine kinase domain by ligation-dependent polymerase chain reaction (LD-PCR). Leukemia. 2008;22(10):1956-1961.
4. Alikian M, Gerrard G, Subramanian PG, et al. BCR-ABL1 kinase domain mutations: methodology and clinical evaluation. Am J Hematol. 2012;87(3):298-304.
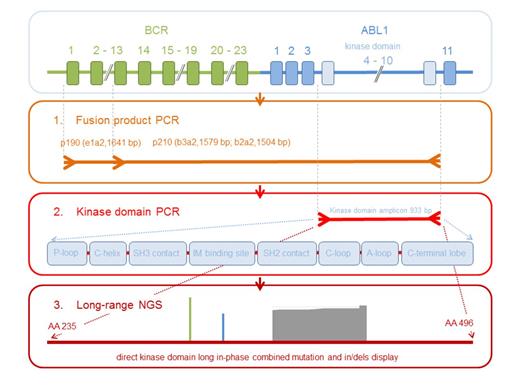
Strategy of long-range NGS analysis for the detection of single and compound mutations, insertions and deletions in the BCR-ABL1 TKD.
Strategy of long-range NGS analysis for the detection of single and compound mutations, insertions and deletions in the BCR-ABL1 TKD.
Valent:Novartis: Honoraria, Research Funding. Lion:Novartis, Bristol-Myers- Squibb, Pfizer: Honoraria, Membership on an entity’s Board of Directors or advisory committees, Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal