Abstract
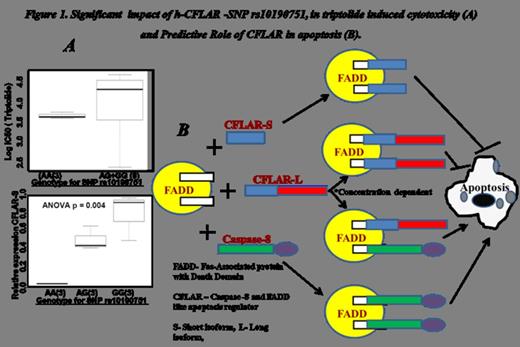
Triptolide is a therapeutic diterpenoid derived from the Chinese herb Tripterygium wilfordii Hook f. It has shown to have immunosuppressive, anti-inflammation and antineoplastic effect Triptolide also serves as a potent inhibitor against different cancer types by regulating/arresting various cell signaling mechanisms such as inducing apoptotic signals by activation of pro-apoptotic proteins, inhibiting NFkB and c-KIT pathways, suppressing the Jak2 transcription, activating MAPK8/JNK signaling and modulating the heat shock responses. Further, as many of the current drugs target p53 mediated pathway to induce growth arrest or apoptosis, dysfunction or mutation in p53 leads to resistance to chemotherapy. In such patients, triptolide provide an edge over other chemotherapeutic drugs, due to its P53 independent mode of action. Genetic subsets for pancreatic cancer cells were found to be significantly correlated with triptolide sensitivity. In recent years, Epstein-Barr-virus transformed lymphoblastoid cell lines (LCLs) that are part of Interenational HapMap project (www.hapMap.org) have been used to study drug response and genetic variation correlation. Genotype data is publically available for these HapMap LCLs, thus allowing genotype-phenotype as well as genome-wide association studies. In the present study, we used HapMap LCLs derived from 60 unrelated subjects with Caucasian ancestry to identify genetic markers that are predictive of cellular sensitivity to triptolide. Our results identified a novel QTL with SNPs on chromosome 2 with significant association with triptolide cellular sensitivity. Detailed analysis of the top 100 highly significant SNPs (observed p<0.0001), identified genes such as CFLAR, PPIl3, Caspase 8, Caspase 10, NFkB and STAT6 that are involved in cell death receptor and apoptotic pathway. As per our findings, the CFLAR topped among all genes which have highest number of SNPs that are significantly associated to triptolide activity as well as showing most promising gene expression correlation It has been reported that CFLAR is a key regulator of apoptosis and chemo resistance in variety of tumors. Hence, our study provides first line of evidence that CFLAR genetic variation may play a crucial role in variety of cancer treatments with triptolide. An interesting SNP, rs10190751 observed in the vicinity of splicing region, is predicted to play a pivotal role in cell cytotoxicity induced by triptolide as it is directly correlated to CFLAR expression (Figure 1). In general, CFLAR is expressed as long (c-FLIPL), short (c-FLIPS) and Raji (c-FLIPR), splice variants in human cells. Variation in rs10190751 (G to A) changes the expression ratio of long to short form (Figure 1), which impacts chemosensitivity of cells towards triptolide. rs10190751 A/A produces a CFLAR-R form instead of CFLAR-S form which increase the sensitivity to triptolide to a significant level. It has been reported in AML that, ratio of CFLAR-L/CFLARs is a potential biomarker for overall survival in an adult patient cohort Overall higher median expression of CFLAR-L has been reported to be significantly correlated with lower overall survival. Our results of CFLAR-knock down in THP1 AML cell line significantly increased sensitivity towards triptolide. In summary, the results of the present study indicates that activation of extrinsic apoptotic pathway and CFLAR protein could be a major signaling cascade for triptolide mediated cytotoxicity. rs10190751, SNP present in CFLAR could be a potential genetic variation marker for a dose dependent effect of triptolide. Overall our results identified novel biomarkers with immense biological significance that can predict response to triptolide and would be of high clinical relevance to patients being treated by triptolide.
Disclosures:
No relevant conflicts of interest to declare.
Author notes
*
Asterisk with author names denotes non-ASH members.
© 2013 by The American Society of Hematology
2013

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal