Key Points
Activated platelets release microRNA miR-223 preferentially through MPs that can be internalized by endothelial cells.
Platelet MP-derived Ago2•microRNA complexes are functional and can regulate endogenous gene expression in recipient endothelial cells.
Abstract
Platelets play a crucial role in the maintenance of hemostasis, as well as in thrombosis. Upon activation, platelets release small membrane-bound microparticles (MPs) containing bioactive proteins and genetic materials from their parental cells that may be transferred to, and exert potent biological effects in, recipient cells of the circulatory system. Platelets have been shown to contain an abundant and diverse array of microRNAs, and platelet-derived MPs are the most abundant microvesicles in the circulation. Here we demonstrate that human platelets activated with thrombin preferentially release their miR-223 content in MPs. These MPs can be internalized by human umbilical vein endothelial cells (HUVEC), leading to the accumulation of platelet-derived miR-223. Platelet MPs contain functional Argonaute 2 (Ago2)•miR-223 complexes that are capable of regulating expression of a reporter gene in recipient HUVEC. Moreover, we demonstrate a role for platelet MP-derived miR-223 in the regulation of 2 endogenous endothelial genes, both at the messenger RNA and protein levels. Our results support a scenario by which platelet MPs may act as intercellular carriers of functional Ago2•microRNA complexes that may exert heterotypic regulation of gene expression in endothelial cells, and possibly other recipient cells of the circulatory system.
Introduction
Platelets are discoid, anucleate cytoplasmic fragments released by bone marrow megakaryocyte precursor cells into the circulation where they play a central role in the maintenance of hemostasis, as well as in thrombosis.1 Platelets are recruited to, and are activated at, sites of damaged vessel walls or atherosclerotic plaques adjacent to the endothelial lining.
We reported that human platelets contain an abundant and diverse array of microRNAs2,3 that may be involved in regulating platelet messenger RNAs (mRNAs),3 protein synthesis,4 and reactivity.5 MicroRNAs are 19- to 24-nucleotide noncoding RNAs6,7 generated by the ribonuclease III Dicer8 and incorporated into effector Argonaute 2 (Ago2) complexes.9 The biological role of microRNAs is linked mainly to their ability to act in concert and mediate sequence-specific regulation (mainly repression10 ) of mRNA translation through recognition of specific binding sites usually located in the 3′ untranslated region (UTR). Predicted to regulate ∼60% of the genes in humans,11 microRNAs may be implicated in the regulation of every cellular process, and changes in their expression and/or function have been associated with human genetic diseases.12,13
Whereas the majority of microRNAs are found intracellularly, a number of microRNAs have also been detected outside of cells, in various body fluids, such as serum or plasma.14 Circulating microRNAs may be found in exosomes,15 shedding vesicles,16 and apoptotic bodies,17 as well as in vesicle-free ribonucleoprotein complexes, in association with Ago218 or high-density lipoproteins (HDL).19
Activated platelets may also release microparticles (MPs), small extracellular vesicles ranging from 0.1 to 1 µm in diameter shed from the cytoplasmic membrane. The MPs derived from platelets are the most abundant cell-derived MP subtype in the circulation20 and may contribute to inflammatory diseases, such as arthritis21 and atherosclerosis.22 In addition to sharing the surface markers of their parental cells, MPs carry a broad variety of cytoplasmic components, including proteins, DNA, and RNA.16 These small lipid vesicles may act, therefore, as intercellular carriers and deliver bioactive proteins and RNAs to recipient cells, including mRNAs23 and small mRNA regulatory microRNAs.24 MPs may, therefore, play an important role as a cargo of genetic information from one cell type to another across the circulatory system,16 including the endothelial cells that line the inner surface of the vasculature.
The relative abundance and diversity of platelet microRNAs,2,3 the propensity of platelets to release MPs upon activation,19 the microRNA content of platelet-derived MPs,25 and the relatively high level of circulating microRNAs originating from platelets26 prompted us to investigate the possible intercellular transfer of platelet microRNAs via MPs and assess their capacity for heterotypic regulation of gene expression in recipient endothelial cells.
Methods
Platelet purification, activation, and MP isolation
Platelets were isolated from venous blood, as previously described,3 harvested by centrifugation at 1000 g for 10 minutes and resuspended at 108 platelets/mL in HEPES-Tyrode buffer (130 mM NaCl, 3 mM KCl, 0.3 mM Na2HPO4, 12 mM NaHCO3, 20 mM HEPES, 5 mM monohydrate d-glucose, 0.5 mM MgCl2, pH 7.4). Platelet activation and MP release were induced upon incubation with 0.1 U/mL thrombin (Sigma-Aldrich, St. Louis, MO) for 15 or 60 minutes at 37°C with gentle agitation. Platelet activation was stopped by the addition of 20 mM final EDTA, and platelets were pelleted by centrifugation at 3,200 g for 10 minutes. The supernatant was centrifuged again to prepare a platelet-free releasate, which was used for MP isolation. MPs were harvested by centrifugation at 20 000 g for 90 minutes at 18°C, and were either resuspended in HEPES-Tyrode buffer for cell coincubations or its RNA extracted by addition of TRIzol (Invitrogen, Carlsbad, CA). The supernatant fraction was collected, snap frozen, and stored at −80°C until analyses.
Flow cytometry
Platelets and the MP fractions were analyzed by flow cytometry to determine their activation status and origin, respectively. Platelets were labeled with CD62P-R-phycoerythrin and CD41a-anti-allophycocyanin (APC) (BD Biosciences, San Jose, CA), and MPs were labeled with CD41a-APC (BD Biosciences). Approximately 10% of unstimulated expressed CD62P at their surface, suggesting that our freshly isolated platelets were minimally activated, as compared with ∼98% of the platelets activated with thrombin (supplementary Figure 1).
MPs (1 µL) were diluted in 100 µL of phosphate buffered saline and incubated in the presence of APC-conjugated mouse IgG (isotype) or APC-conjugated anti-human CD41a (BD Biosciences) for 30 minutes. Then the samples were diluted to 500 µL in phosphate buffered saline and analyzed cytofluorometrically using a forward-scattered light coupled to a photomultiplier tube (PMT) option (forward-scattered PMT) (BD Biosciences) mounted on a Canto II SORP flow cytometer (BD Biosciences). The cytometer was calibrated before all data acquisitions using BD cytometer setup and tracking beads (BD Biosciences).
Cell culture
HUVEC (Stem Cell Technologies, Vancouver, BC, Canada) were cultured in endothelial growth medium (Lonza, Basel, Switzerland) supplemented with bovine brain extract (Lonza) and maintained at 37°C under 5% CO2. For all experiments, HUVEC were used between passages 2 to 6. For transfection, 5 × 105 cells were transfected by nucleofection on the Nucleofector II apparatus (Lonza) using the AMAXA HUVEC Nucleofector kit (Lonza). Twenty-four hours later, the culture medium was changed and the MPs were added for up to 48 hours of coincubation.
Production of labeled MP and HUVEC-MP coincubation assays
Isolated human platelets (108 platelets/mL in HEPES-Tyrode buffer) were incubated with 1 µM CellTracker Orange CMTMR (5-[and-6]-[([4-Chloromethyl]Benzoyl)Amino]Tetramethylrhodamine) (Invitrogen) for 15 minutes at 37°C in darkness, prior to platelet activation with 0.1 U/mL thrombin. The labeled platelet-derived MPs were recovered by centrifugation, resuspended in HEPES-Tyrode buffer, and counted by flow cytometry. HUVEC were incubated with fluorescent MPs at a ratio of 1:100 (HUVEC:MPs) for up to 48 hours at 37°C under 5% CO2. MP internalization was evaluated by confocal microscopy analysis with a spinning disc confocal microscope using a ×63 objective (Quorum Spinning Disc Wave FX, Quorum Technologies, Guelph, ON, Canada). Up to 24 images, corresponding to as many 0.5 µm-thick layers, were acquired using the Volocity software (PerkinElmer, Waltham, MA). Single representative images of the central layers are shown.
RNA extraction, microRNA quantification, and gene expression studies
Total RNA was extracted from platelet-derived MPs and HUVEC using TRIzol reagent (Invitrogen), and from the supernatant fraction using mirVana PARIS kit (Ambion, Austin, TX). Reverse transcription reactions were performed with 1 µg total RNA using HiFlex miSCRIPT RTII kit (Qiagen, Hilden, Germany) after DNase I treatment (Invitrogen). Mature miR-223 and 2 selected mRNAs were detected by quantitative PCR (qPCR) using miScript Primer Assay kit and SYBR Green (Qiagen). Small nuclear RNA U6 (RNU6) (for miR-223) and glyceraldehyde-3-phosphate dehydrogenase (for mRNAs) were used as reference genes for relative quantitation using the 2^-ΔΔCt method.27 The sequence of the oligonucleotides used for qPCR quantitation of selected endothelial mRNAs are provided in supplementary Table 1.
Immunoprecipitation and functional assay of Ago2•microRNA complexes
MPs derived from activated platelets were lysed in RNA immunoprecipitation lysis buffer, and the lysates cleared by centrifugation prior to immunoprecipitation using protein G-agarose beads (Roche Applied Science, Penzberg, Germany) conjugated with anti-Ago2 antibody (clone 2E12-1C9, Abnova) or isotypic IgG control (anti-FLAG; Sigma-Aldrich), as previously described.3,28 Ago2-associated miR-223 was isolated by phenol/chloroform extraction and ethanol precipitation, reverse transcribed with the miScript II RT kit (Qiagen), and analyzed by qPCR using hsa-miR-223 miScript Primer Assay (Qiagen).
Ago2⋅miR-223 function was evaluated in RNA-induced silencing complex activity assays, as previously described.3
Reporter gene activity assays
Reporter gene activity assays were performed essentially as previously described.2,29,30 A miR-223 reporter construct was created by inserting a sequence complementary to hsa-miR-223 in the Xba1 site of pRL-CMV vector (Promega, Madison, WI), downstream of the Renilla luciferase (Rluc) reporter gene. pRL-CMV-3′UTR Ephrin A1 (EFNA1) and pRL-CMV-3′UTR F-box/WD repeat-containing protein 7 (FBXW7) constructs were engineered by amplifying and cloning their 3′UTR element downstream of the Rluc reporter gene in pRL-CMV vector. All the constructs were verified by DNA sequencing. The pGL4.51 vector (Promega) expressing Firefly luciferase (Fluc) was used as a normalization control. Both pRL-CMV and pGL4.51 constructs were co-transfected in HUVEC 24 hours prior to incubation with MPs for up to 48 hours. Rluc and Fluc activities were measured with Dual Glo luciferase reagents (Promega) using a luminometer (Dynex Technologies, Chantilly, VA).
Western blot analysis
Protein extracts were analyzed by 10% (wt/vol) sodium dodecyl sulfate polyacrylamide gel electrophoresis and immunoblotting using anti-Ago2 (Abnova, Taipei City, Taiwan), anti-EFNA1 (Abcam, Cambridge, UK), anti-FBXW7 (Invitrogen), and anti-β-actin (AC-40; Sigma-Aldrich) antibodies, followed by enhanced chemiluminescence detection and densitometric analyses, as previously described.3,29
Blood collection from healthy volunteers (adult Caucasians of both sexes from the immediate region of Quebec City) was approved by our institutional human ethics committee. The participants provided their written informed consent to participate in this study in accordance with the Declaration of Helsinki, as approved by our institutional human ethics committee.
Results
Activated platelets release MPs that contain miR-223
Flow cytometry analyses of MPs purified from resting or thrombin-activated platelets unveiled a predominant population of MPs, approximately 100 to 400 nm in diameter, which together with platelet glycoprotein CD41a surface expression (Figure 1A) confirmed the platelet origin of these MPs.31
Activated platelets release MPs that contain miR-223. (A-C) Human platelets were activated with thrombin (0.1 U/mL) for 15 or 60 minutes, and the MPs released were isolated by ultracentrifugation. (A) Representative flow cytometry analyses of the MP population derived from platelets activated with thrombin for 60 minutes. Because of the size of heterogeneity of the MPs,49 fluorescent Sky Blue microspheres, ranging from 90 nm to 3200 nm in diameter (Spherotech, Lake Forest, IL), were used to calibrate our flow cytometer and estimate the size of the MPs. The CD41a+ events were portrayed as forward-scattered light (FSC) and side-scattered light (SSC) PMT graph using the BD FACSDiva software. (B) MPs were counted by flow cytometry by an FSC coupled to a PMT. Results are expressed as the mean (± standard error of the mean [SEM]) fold changes vs unstimulated platelets, used as a reference (n = 3 experiments). (C) The MP and supernatant fractions of thrombin-activated platelets were isolated and analyzed for their content in miR-223 by qPCR. Results were normalized by the 2^-ΔΔCt method, using RNU6 as a reference,27 and expressed as the mean (±SEM) fold changes vs unstimulated platelets (n = 3 experiments). Similar results were obtained by monitoring 4 additional microRNAs (data not shown). *P < .05 vs baseline (Student t test). SSC, side-scattered light.
Activated platelets release MPs that contain miR-223. (A-C) Human platelets were activated with thrombin (0.1 U/mL) for 15 or 60 minutes, and the MPs released were isolated by ultracentrifugation. (A) Representative flow cytometry analyses of the MP population derived from platelets activated with thrombin for 60 minutes. Because of the size of heterogeneity of the MPs,49 fluorescent Sky Blue microspheres, ranging from 90 nm to 3200 nm in diameter (Spherotech, Lake Forest, IL), were used to calibrate our flow cytometer and estimate the size of the MPs. The CD41a+ events were portrayed as forward-scattered light (FSC) and side-scattered light (SSC) PMT graph using the BD FACSDiva software. (B) MPs were counted by flow cytometry by an FSC coupled to a PMT. Results are expressed as the mean (± standard error of the mean [SEM]) fold changes vs unstimulated platelets, used as a reference (n = 3 experiments). (C) The MP and supernatant fractions of thrombin-activated platelets were isolated and analyzed for their content in miR-223 by qPCR. Results were normalized by the 2^-ΔΔCt method, using RNU6 as a reference,27 and expressed as the mean (±SEM) fold changes vs unstimulated platelets (n = 3 experiments). Similar results were obtained by monitoring 4 additional microRNAs (data not shown). *P < .05 vs baseline (Student t test). SSC, side-scattered light.
The number of MPs released from platelets significantly increased by 2.1-fold as early as 15 minutes after stimulation with thrombin (Figure 1B). This increase reached up to 7.3-fold after 60 minutes. Knowing that platelets can release microRNAs25,26 and that microvesicles may contain microRNAs,16 we assessed whether activated platelets release miR-223 (one of the most abundant platelet microRNA)2,3 either directly into the supernatant or through MPs. Little or no changes were observed in supernatant microRNA levels (Figure 1C). However, we observed a significant ∼60-fold increase in MP miR-223 levels 60 minutes after platelet activation with thrombin, as compared with baseline (Figure 1C), suggesting that thrombin-activated platelets may preferentially release miR-223 through MPs.
Platelet-derived MPs contain functional Ago2•miR-223 effector complexes
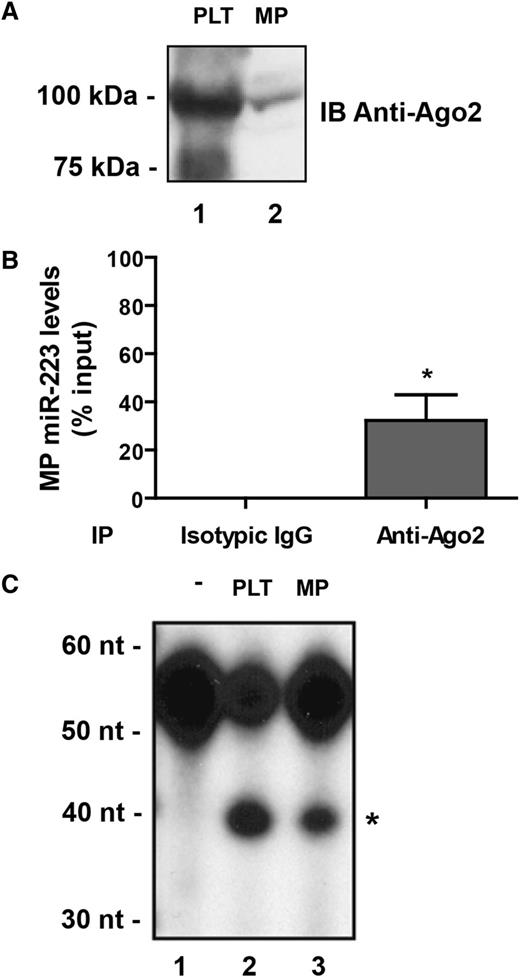
MicroRNAs are known to guide effector complexes containing Ago proteins, such as Ago2,9 for the regulation of specific mRNAs through translational repression, mRNA destabilization, or a combination of the these.11 Immunoblot analysis of platelet-derived MP protein extracts revealed the presence of Ago2 proteins (Figure 2A, lane 2), which together with the presence of MP miR-223 prompted us to verify if Ago2 and miR-223 exist in MPs in the form of a complex.
Platelet-derived MPs contain functional Ago2•miR-223 effector complexes. (A-C) Human platelets (PLT) were activated with thrombin (0.1 U/mL) for 60 minutes, and the MP fraction was isolated by ultracentrifugation. Unstimulated platelets served as the control.3 (A) The presence of Ago2 was assessed by immunoblot (IB) analysis using an anti-Ago2 antibody. (B) Protein extracts derived from the MP fraction were subjected to immunoprecipitation using anti-Ago2 antibody, followed by quantitative miR-223 detection by qPCR. Results were normalized by the 2^-ΔΔCt method, using RNU6 as a reference,27 and expressed as a percentage of the input (mean ± SEM; n = 4 experiments). *P < .05 vs normal isotypic IgG, which was used as an IP control (Student t test). (C) The supernatant (S100) (50 µg proteins) fraction of protein extracts derived from PLT or the MP fraction of thrombin-activated platelets were subjected to RNA target cleavage assays, using a 5′ end, 32P-labeled miR-223 RNA sensor. RNA was isolated by phenol/chloroform extraction and ethanol precipitation, separated by denaturing 8% polyacrylamide gel electrophoresis PAGE/7 M urea and visualized by autoradiography (n = 2 experiments). *The 39-nucleotide RNA product expected from Ago2-mediated endonucleolytic cleavage of the sensor. -, uncleaved 32P-labeled miR-223 RNA sensor.
Platelet-derived MPs contain functional Ago2•miR-223 effector complexes. (A-C) Human platelets (PLT) were activated with thrombin (0.1 U/mL) for 60 minutes, and the MP fraction was isolated by ultracentrifugation. Unstimulated platelets served as the control.3 (A) The presence of Ago2 was assessed by immunoblot (IB) analysis using an anti-Ago2 antibody. (B) Protein extracts derived from the MP fraction were subjected to immunoprecipitation using anti-Ago2 antibody, followed by quantitative miR-223 detection by qPCR. Results were normalized by the 2^-ΔΔCt method, using RNU6 as a reference,27 and expressed as a percentage of the input (mean ± SEM; n = 4 experiments). *P < .05 vs normal isotypic IgG, which was used as an IP control (Student t test). (C) The supernatant (S100) (50 µg proteins) fraction of protein extracts derived from PLT or the MP fraction of thrombin-activated platelets were subjected to RNA target cleavage assays, using a 5′ end, 32P-labeled miR-223 RNA sensor. RNA was isolated by phenol/chloroform extraction and ethanol precipitation, separated by denaturing 8% polyacrylamide gel electrophoresis PAGE/7 M urea and visualized by autoradiography (n = 2 experiments). *The 39-nucleotide RNA product expected from Ago2-mediated endonucleolytic cleavage of the sensor. -, uncleaved 32P-labeled miR-223 RNA sensor.
With the aim to subsequently study the role of platelet-derived MP microRNAs in regulating endothelial cell gene expression, we selected a microRNA that is highly abundant in platelets while being expressed at very low levels in endothelial cells (ie, miR-223).3,24,25,32 Ago2 immunoprecipitation followed by qPCR detection of miR-223 unveiled the existence of an Ago2⋅miR-223 complex in MPs released from thrombin-activated platelets (Figure 2B). The levels of miR-223 associated with MP Ago2 proteins were enriched by more than 300-fold compared with an isotypic control IgG (anti-FLAG antibody) (supplementary Figure 2).
To determine whether MP Ago2⋅miR-223 complexes are functional, we performed RNA-induced silencing complex activity assays in which MP or platelet protein extracts were incubated in the presence of a 32P-labeled RNA sensor bearing a sequence complementary to hsa-miR-223. We observed the cleavage of the RNA sensor into a 39-nucleotide RNA product (Figure 2C), supporting the functionality of Ago2⋅miR-223 complexes present in MPs released from activated platelets. Together, these results suggest that platelet-derived MPs can participate to intercellular communication by delivering functional Ago2⋅microRNA complexes to recipient cells.
Platelet-derived MPs are internalized by HUVEC
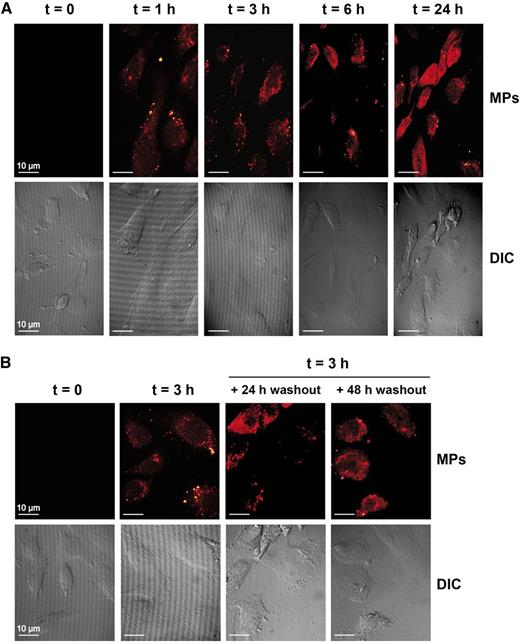
First, we used confocal fluorescence microscopy to determine if platelet-derived MPs could be internalized by HUVEC. We observed the presence of well-defined punctate signals in the cytoplasm of HUVEC exposed to fluorescently labeled platelet-derived MPs for 1 hour (Figure 3A, upper panel, second from left), which is consistent with the internalization of platelet-derived MPs by HUVEC. The fluorescent signal persisted for at least 24 hours, raising the issue as to whether this is due to the continuous uptake of MPs or the stability of MPs in HUVEC. To verify that possibility, we coincubated HUVEC with labeled MPs for 3 hours to allow MP internalization before washing the cells, changing the medium, and prolonging the incubation period to 48 hours. We observed that the cytoplasmic punctate staining of HUVEC, conferred by the fluorescent MPs, persisted for 48 hours in the absence of exogenous MPs (Figure 3B, upper panels). We noticed that the fluorescence seemed to have diffused over time (Figures 3A-B), which is in accordance with the release of the MP content in the HUVEC cytoplasm.
Platelet-derived MPs are internalized by HUVEC. (A-B) HUVEC were incubated for the indicated periods of time (t), with fluorescent-labeled MPs at an HUVEC:MPs ratio of 1:100, and an MP uptake was visualized by confocal microscopy (×63 objective) (upper panels). Cell morphology was visualized using differential interference contrast (DIC) (lower panels). (B) HUVEC were incubated with fluorescently labeled MPs for 3 hours, and MP uptake was visualized by confocal microscopy after a 24- or 48-hour washout period (upper panels). The images are representative of 3 independent experiments.
Platelet-derived MPs are internalized by HUVEC. (A-B) HUVEC were incubated for the indicated periods of time (t), with fluorescent-labeled MPs at an HUVEC:MPs ratio of 1:100, and an MP uptake was visualized by confocal microscopy (×63 objective) (upper panels). Cell morphology was visualized using differential interference contrast (DIC) (lower panels). (B) HUVEC were incubated with fluorescently labeled MPs for 3 hours, and MP uptake was visualized by confocal microscopy after a 24- or 48-hour washout period (upper panels). The images are representative of 3 independent experiments.
Platelet-derived MPs can deliver functional Ago2•miR-223 effector complexes to HUVEC
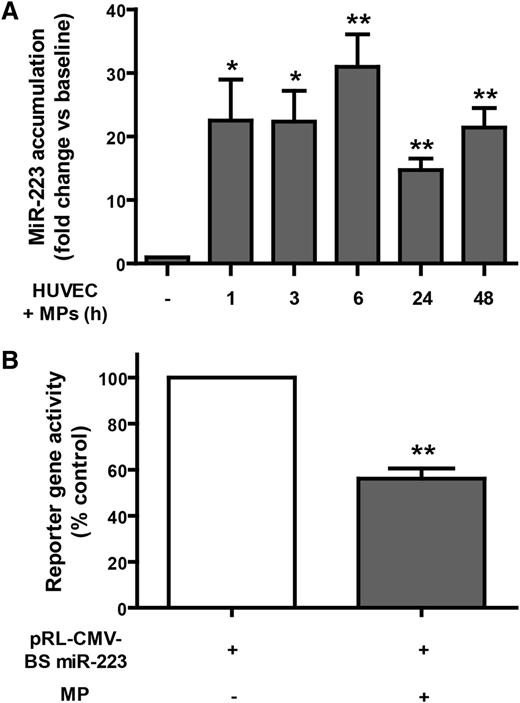
Knowing that platelet-derived MPs contain an appreciable amount of micoRNAs,24,25 we wanted to determine if MPs could deliver their microRNA content to HUVEC after their internalization. Focusing on miR-223, which is highly abundant in platelets3 but weakly expressed in HUVEC,32 we observed a 22-fold increase in HUVEC miR-223 levels as early as 1 hour after incubation with platelet-derived MPs (Figure 4A). This significant enrichment in HUVEC miR-223 levels persisted for up to 48 hours, compared with HUVEC incubated without MPs. The marked increase and persistence of elevated miR-223 levels in HUVEC may provide the conditions and time window necessary for platelet-derived Ago2⋅miR-223 to regulate HUVEC gene expression.
Platelet-derived MPs can deliver functional Ago2•microRNA effector complexes to HUVEC. (A) HUVEC were incubated with platelet-derived MPs for up to 48 hours, and miR-223 accumulation in HUVEC was quantitated by qPCR. Results were normalized by the 2^-ΔΔCt method, using RNU6 as a reference,27 and expressed as the mean (±SEM) fold changes vs baseline (n = 5 experiments). (B) HUVEC transiently expressing a Rluc reporter gene, harboring a binding site complementary to miR-223 (pRL-CMV-BS miR-223), were incubated (or not; control) with MPs derived from thrombin-activated platelets for 48 hours prior to luciferase activity measurements. Results were normalized on Fluc activity, and expressed as mean (±SEM) percentage of control (n = 4 experiments). *P < .05; **P < .01 vs baseline or control (Student t test). pRL-CMV, Renilla luciferase–cytomegalovirus (CMV) plasmid; BS miR-223, Binding site for miR-223.
Platelet-derived MPs can deliver functional Ago2•microRNA effector complexes to HUVEC. (A) HUVEC were incubated with platelet-derived MPs for up to 48 hours, and miR-223 accumulation in HUVEC was quantitated by qPCR. Results were normalized by the 2^-ΔΔCt method, using RNU6 as a reference,27 and expressed as the mean (±SEM) fold changes vs baseline (n = 5 experiments). (B) HUVEC transiently expressing a Rluc reporter gene, harboring a binding site complementary to miR-223 (pRL-CMV-BS miR-223), were incubated (or not; control) with MPs derived from thrombin-activated platelets for 48 hours prior to luciferase activity measurements. Results were normalized on Fluc activity, and expressed as mean (±SEM) percentage of control (n = 4 experiments). *P < .05; **P < .01 vs baseline or control (Student t test). pRL-CMV, Renilla luciferase–cytomegalovirus (CMV) plasmid; BS miR-223, Binding site for miR-223.
To verify this, we transiently transfected HUVEC with a reporter gene construct, in which Rluc was placed under the control of a miR-223 binding site, prior to incubation with platelet-derived MPs. We observed a significant 44% decrease in HUVEC reporter gene activity induced by coincubation with MPs (Figure 4B). These results indicate that platelet MP-derived Ago2⋅miR-223 complexes can target specific mRNAs and exert gene regulatory effects in recipient endothelial cells.
Platelet MP-derived miR-223 can regulate HUVEC gene expression at the mRNA level
To investigate whether MP miR-223 derived from platelets can regulate endogenous HUVEC gene expression, we searched for potential endothelial mRNA targets of platelet miR-223, and identified 2 mRNA candidates: FBXW7, an onco-suppressor protein and validated mRNA target of miR-22333 ; and EFNA1, a glycosylphosphatidyl inositol-anchored receptor tyrosine kinase ligand.34 FBXW7 and EFNA1 mRNAs harbor 4 and 1 conserved miR-223 binding sites in their 3′UTR, respectively (supplementary Figure 3).35
Using a reporter gene-based assay in HUVEC, we observed a significant 47% and 31% downregulation of Rluc reporter gene activity induced upon coincubation with MPs and conferred by the 3′UTR of FBXW7 (Figure 5A, left panel) and EFNA1 (Figure 5A, right panel) mRNAs, respectively. These results indicate that platelet-derived MPs can modulate gene expression through regulatory elements located in the 3′UTR of 2 selected endothelial mRNAs.
Platelet MP-derived miR-223 can regulate HUVEC gene expression at the mRNA level. (A) HUVEC transiently expressing a Rluc reporter gene, harboring the 3′UTR of FBXW7 (pRL-CMV-3′UTR FBXW7) (left panel) or EFNA1 mRNA (pRL-CMV-3′UTR EFNA1) (right panel), were incubated (or not; control) with MPs derived from thrombin-activated platelets for 48 hours prior to luciferase activity measurements. Results were normalized on Fluc activity, and expressed as mean (±SEM) percentage of control (n = 3 experiments). (B-C) HUVEC transiently expressing a miR-223 sponge (pRL-CMV-BS miR-223 vector; n = 3 experiments) (C), or not (n = 5 experiments) (B), were incubated with platelet-derived MPs for up to 48 hours, and FBXW7 (left panels) and EFNA1 (right panels) mRNA levels were quantitated by qPCR. Results were normalized by the 2^-ΔΔCt method, using glyceraldehyde-3-phosphate dehydrogenase mRNA as a reference, and expressed as mean (±SEM) fold changes vs baseline. *P < .05; **P < .01; ***P < .0001 vs control or baseline (Student t test).
Platelet MP-derived miR-223 can regulate HUVEC gene expression at the mRNA level. (A) HUVEC transiently expressing a Rluc reporter gene, harboring the 3′UTR of FBXW7 (pRL-CMV-3′UTR FBXW7) (left panel) or EFNA1 mRNA (pRL-CMV-3′UTR EFNA1) (right panel), were incubated (or not; control) with MPs derived from thrombin-activated platelets for 48 hours prior to luciferase activity measurements. Results were normalized on Fluc activity, and expressed as mean (±SEM) percentage of control (n = 3 experiments). (B-C) HUVEC transiently expressing a miR-223 sponge (pRL-CMV-BS miR-223 vector; n = 3 experiments) (C), or not (n = 5 experiments) (B), were incubated with platelet-derived MPs for up to 48 hours, and FBXW7 (left panels) and EFNA1 (right panels) mRNA levels were quantitated by qPCR. Results were normalized by the 2^-ΔΔCt method, using glyceraldehyde-3-phosphate dehydrogenase mRNA as a reference, and expressed as mean (±SEM) fold changes vs baseline. *P < .05; **P < .01; ***P < .0001 vs control or baseline (Student t test).
We observed a significant downregulation of endogenous FBXW7 and EFNA1 mRNA levels that reached a maximum of 51% (Figure 5B, left panel) and 28% (Figure 5B, right panel), respectively, within 6 hours of exposure to MPs. To ascertain that these effects are mediated by platelet MP-derived miR-223, we transiently transfected HUVEC, with a vector expressing a mRNA bearing a sequence complementary to miR-223 and acting as a miR-223 sponge, prior to incubation with MPs. As shown in Figure 5C, the miR-223 sponge neutralized the down-regulatory effects induced by platelet MPs on the endogenous FBXW7 (Figure 5C, left panel) and EFNA1 (Figure 5C, right panel) mRNA levels, and even enhanced EFNA1 mRNA levels above baseline. This latter observation may be due to the sequestration of endothelial miR-223, which may regulate endogenous FBXW7 expression to a greater extent than EFNA1. These findings support a role for platelet MP-derived miR-223 in regulating FBXW7 and EFNA1 expression at the mRNA level.
Platelet MP-derived miR-223 can regulate HUVEC gene expression at the protein level
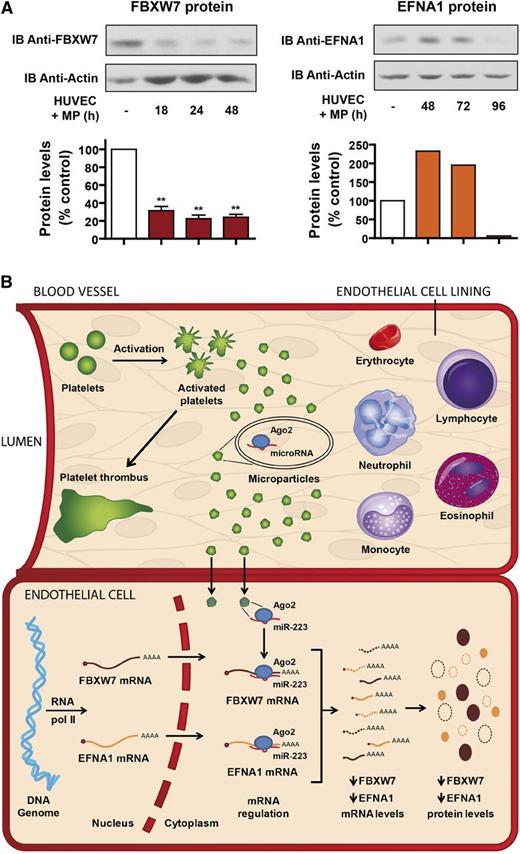
The mRNA regulatory effects of platelet MP-derived miR-223 could be translated at the protein levels, as we observed a marked decrease in HUVEC FBXW7 protein levels as early as 18 hours after exposure to platelet-derived MPs (Figure 6A, left panel). This effect persisted for up to 48 hours, suggesting that platelet MP-derived miR-223 may influence endothelial gene expression for extended periods of time. HUVEC EFNA1 protein levels were also decreased by platelet-derived MPs, but only after 96 hours of coincubation (Figure 6A, right panel), which is probably due to a slower turnover of cellular EFNA1 proteins compared with FBXW7.
Platelet MP-derived miR-223 can regulate HUVEC gene expression at the protein level. (A) Protein extracts prepared from HUVEC, incubated or not with MPs derived from thrombin-activated platelets for up to 48 or 96 hours, were analyzed by immunoblotting (IB) using anti-F-box and WD-40 domain protein 7 (anti-FBXW7) (left upper panel; n = 3 experiments), anti-EFNA1 (right upper panel; n = 1 experiment) or anti-actin (lower panels) antibody, which was used as a loading control. The data were analyzed by densitometry and expressed as a percentage of control. (B) Proposed model for the intercellular transfer of functional Ago2•microRNA complexes between activated platelets and endothelial cells through the release of MPs. RNA pol II, RNA polymerase II.
Platelet MP-derived miR-223 can regulate HUVEC gene expression at the protein level. (A) Protein extracts prepared from HUVEC, incubated or not with MPs derived from thrombin-activated platelets for up to 48 or 96 hours, were analyzed by immunoblotting (IB) using anti-F-box and WD-40 domain protein 7 (anti-FBXW7) (left upper panel; n = 3 experiments), anti-EFNA1 (right upper panel; n = 1 experiment) or anti-actin (lower panels) antibody, which was used as a loading control. The data were analyzed by densitometry and expressed as a percentage of control. (B) Proposed model for the intercellular transfer of functional Ago2•microRNA complexes between activated platelets and endothelial cells through the release of MPs. RNA pol II, RNA polymerase II.
Together, these results are consistent with a scenario by which activated platelets may deliver, through the release of MPs, mRNA regulatory Ago2⋅microRNA complexes to other cells of the cardiovascular system and regulate expression of endogenous genes in recipient cells, such as endothelial cells (Figure 6B).
Discussion
Although recent studies tend to support a mRNA regulatory role for platelet microRNAs4 and Ago2⋅microRNA complexes3 in platelet biology,4,5 the definite proof is still being sought, because of the challenge and intrinsic limitations associated with working with primary human platelets, including their relative refractoriness to transfection.3 The propensity of platelets to release MPs upon activation,21 the microRNA content of circulating MPs derived from platelets,25 and the capacity of MPs to transfer their content to recipient cells16 made it tempting to investigate a possible role of platelet microRNAs outside of platelets.
The pool of circulating microRNAs is composed of vesicle-associated microRNAs, as well as protein-bound microRNAs that originate from different cell types, including platelets,26 from which the most abundant MP subtype in the circulation derive.20 In the present study, we found that thrombin-activated platelets release a majority of their miR-223 content in MPs, whereas minute amounts seem to be released directly into the supernatant. These findings are supported by Diehl et al,24 who reported that a prominent amount of plasma microRNAs is associated to MPs. The increase in MP miR-223 levels was proportionally superior to, and correlated with, the number of MPs released by thrombin-activated platelets (∼60-fold versus 7.3-fold). These results suggest that thrombin may induce the release of platelet MPs enriched in miR-223, compared with MPs produced spontaneously by resting platelets. However, MPs may not have the monopoly of intercellular microRNA transfer, as circulating microRNAs may also be delivered to target cells via HDL.19 Vickers et al19 reported that miR-223 complexed with HDL can be efficiently transferred to cocultured hepatocytes and influence the expression of miR-223 targets in recipient cells. Our results suggest that activated platelets may not contribute to a significant extent to the pool of vesicle-free forms of circulating microRNAs that are either associated with Ago218 or HDL.19
Platelets may also release microvesicles smaller than MPs, called exosomes (40 to 100 nm in diameter),36 that may not have sedimented with MPs upon centrifugation at 20 000 g. Although miR-223 of exosomal origin could have contributed to the miR-223 signal detected in the supernatant fraction, its overall contribution to the pool of miR-223 released from activated platelets is almost negligible compared with MPs. The caveat has to be taken into account that the profile and mode of microRNA release from platelets may depend on the nature of the agonist(s), stimulatory, and/or shear conditions.
Released upon activation or apoptosis of almost every blood cell type,37 MPs have been associated with a variety of pathologies, including arthritis21 and atherosclerosis,22 as well as tumor progression, angiogenesis, and metastasis.38 This may be explained by the properties of MPs to enhance vascular permeability,39 to promote inflammation,21,31 and to act as a procoagulant.40 Although cytokines may account for most of these biological effects,22,37 it would not be prudent to dismiss a possible role for bioactive molecules, other than cytokines such as microRNAs, in MP-mediated effects in the circulatory system.
MPs and other vesicles, such as exosomes and even apoptotic bodies, can act as carriers and mediate the horizontal transfer of proteins and RNAs between cells. For instance, mRNAs contained in murine mast cell-derived exosomes may be transferred to human mast cells and induce mouse protein expression.15 Platelet-like particles were also shown to transfer their mRNA content to recipient vascular cells.23 Similarly, Zernecke et al17 demonstrated that miR-126 derived from endothelial apoptotic bodies could be efficiently delivered to neighboring endothelial cells and have an atheroprotective role in the mouse. By demonstrating a role for platelet MPs in miR-223 delivery to recipient cells of endothelial origin where it can modulate expression of endogenous genes, our study highlights the relative complexity, efficiency, and functional importance of intercellular communications across the circulatory system. These communications are even more complex, considering the diversity of platelet microRNA sequences,2 as well as the number of cell types capable of exchange (ie, releasing, internalizing, and integrating, genetic materials or information) through different modes of intercellular transfer. In that context, a microvesicular carrier harboring specific surface proteins may confer a certain degree of specificity and provide a means of delivering platelet microRNAs to specific cells. In view of our findings, it may be reasonable to speculate that MPs released by activated platelets may communicate this information, and modulate, indeed adapt, the responsiveness of the vasculature, accordingly. The abundance of circulating MPs may underlie their relative importance in mediating normal regulatory functions and cell-to-cell communications/coordinations, whose dysfunction may contribute to pathophysiological conditions.21
MPs of various origins contain microRNAs and can be internalized by recipient cells.24,25,41 In this study, we report the internalization by HUVEC of platelet MP-derived miR-223, which maintains its functionality and regulates endothelial expression of 2 of its validated mRNA targets (ie, FBXW7 and EFNA1). EFNA1 is a marker of liver cancer,42 whereas FBXW7 was unveiled as a general tumor suppressor in human cancer.43 Both genes are (down)regulated by miR-223 at the mRNA and protein levels. Mechanistically, MP-derived miR-223 may regulate FBXW7 and EFNA1 gene expression, either through mRNA destabilization, inhibition of mRNA translation initiation, or both.11 The fact that downregulation of mRNA levels preceded that of protein levels militates in favor of miR-223-mediated destabilization of these 2 endothelial mRNAs.
Although our results support the involvement of platelet-derived Ago2⋅microRNA complexes in regulating endothelial mRNA translation, we cannot exclude the possibility that MP microRNAs, free of Ago2 proteins, may integrate the microRNA machinery of endothelial cells to mediate their mRNA regulatory effects. In addition, whether platelet-derived microRNAs can mediate epigenetic effects in recipient cells should also be considered.
MiR-223 may be implicated in cell proliferation,44 in osteoclast, erythroid or megakaryocyte differentiation,45,46 and in cancer.47 Therefore, the delivery of platelet miR-223 to other cell types via MPs may have important implications that can only be magnified by the plurality of microRNA targets and the diversity of the microRNA population released through MPs. Such intercellular exchanges of microRNAs mediated by platelet-derived MPs may contribute to the gene expression programming of recipient cells and to the conditioning of the circulatory system under specific health and disease conditions associated with platelet activation.
The in vivo relevance of this process has gained support from a recent study performed by Gidlöf et al,48 in which the authors reported the (down)regulation of intercellular adhesion molecule 1 gene expression in cultured human microvascular endothelial cell–1 cells exposed to a microRNA (ie, miR-320b) that is released upon platelet activation and is found to be depleted in platelet-containing thrombi aspirated from patients with ST-elevation myocardial infarction. The process that we documented may be manipulated and eventually lead to the development of new therapeutic modalities aimed to improve the circulatory function of cardiovascular patients.
The online version of this article contains a data supplement.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
A.-C.D. was supported by a studentship from the Canadian Arthritis Network. E.B. is a New Investigator of the Canadian Arthritis Network and Junior 1 Scholar from the Fonds de recherche du Québec – Santé.
This work was supported by a grant from the Canadian Blood Services/Canadian Institutes of Health Research (CIHR) Blood Utilization and Conservation Initiative via Health Canada (286777) (P.P.) and by a grant from The Arthritis Society/CIHR (244472) (E.B.).
Authorship
Contribution: P.P. conceived and coordinated the project; B.L. led the project; B.L, E.B., and P.P. designed and planned the experiments; B.L., A.C., H.P., A.-C.D. and N.C. performed the experiments and analyzed the data; B.L., A.C., H.P., A.-C.D., N.C., E.B., and P.P. commented on and edited the manuscript; and B.L and P.P. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Patrick Provost, CHUQ Research Center/CHUL, 2705 Blvd Laurier, Room T1-49, Quebec, QC G1V 4G2 Canada; e-mail: patrick.provost@crchul.ulaval.ca.

![Figure 1. Activated platelets release MPs that contain miR-223. (A-C) Human platelets were activated with thrombin (0.1 U/mL) for 15 or 60 minutes, and the MPs released were isolated by ultracentrifugation. (A) Representative flow cytometry analyses of the MP population derived from platelets activated with thrombin for 60 minutes. Because of the size of heterogeneity of the MPs,49 fluorescent Sky Blue microspheres, ranging from 90 nm to 3200 nm in diameter (Spherotech, Lake Forest, IL), were used to calibrate our flow cytometer and estimate the size of the MPs. The CD41a+ events were portrayed as forward-scattered light (FSC) and side-scattered light (SSC) PMT graph using the BD FACSDiva software. (B) MPs were counted by flow cytometry by an FSC coupled to a PMT. Results are expressed as the mean (± standard error of the mean [SEM]) fold changes vs unstimulated platelets, used as a reference (n = 3 experiments). (C) The MP and supernatant fractions of thrombin-activated platelets were isolated and analyzed for their content in miR-223 by qPCR. Results were normalized by the 2^-ΔΔCt method, using RNU6 as a reference,27 and expressed as the mean (±SEM) fold changes vs unstimulated platelets (n = 3 experiments). Similar results were obtained by monitoring 4 additional microRNAs (data not shown). *P < .05 vs baseline (Student t test). SSC, side-scattered light.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/122/2/10.1182_blood-2013-03-492801/4/m_253f1.jpeg?Expires=1764972902&Signature=vpMV2zMscBRVZ9KKx2b5TbyjlLXnWWJdP4yAWX6O8azEXoFRtLdQteeox9mKc8a0xLd9uaz8Aa4~a5xz6ENygIU266TPLGQb1TBclc7XFD4MvOXXPzfYGAkOTt~jIiFPWinVmEMt1e8nV6uBHwE1Nw9-xOchv63cAG7MnD5VLN8wP64aTiR51TbyJaodrv-JRz1-wv2U~5zZeLbfVJCwjVcRry44n~64bi4KeLTIX8HSmhhTFwz-BvUqLkIiy8spRGwbu9P9BxxSndKYoc2VrmFYxA-mzqIpd6cpUfwv3pCBFWBN2Du30NCNK6bxszz-kzB8dSJQCjZXi5ZklQP7oA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)