In this issue of Blood, Yoshida et al present a detailed evaluation of the transcriptional regulation of Ikzf1 (Ikaros) expression during hematopoiesis.1
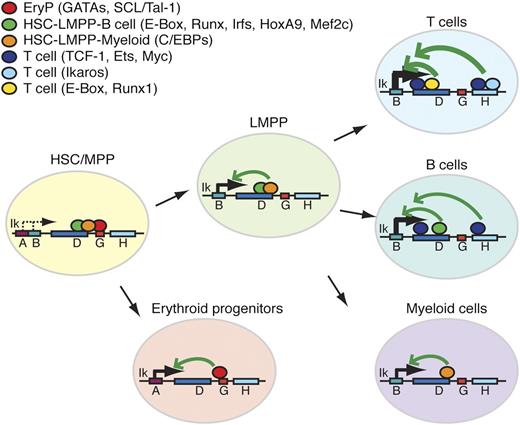
Ikzf1 enhancer-promoter interactions during hematopoiesis. Lineage and/or stage associated transcription factor binding at enhancers of the Ikzf1 locus. Lineage-specific transcription factors at these sites are depicted as color-coded circles. Arrows indicate potential interactions between cell type–specific enhancers and promoters supporting Ikzf1 expression at appropriate developmental stages.
Ikzf1 enhancer-promoter interactions during hematopoiesis. Lineage and/or stage associated transcription factor binding at enhancers of the Ikzf1 locus. Lineage-specific transcription factors at these sites are depicted as color-coded circles. Arrows indicate potential interactions between cell type–specific enhancers and promoters supporting Ikzf1 expression at appropriate developmental stages.
Ikzf1 (Ikaros) functions as a master regulator of hematopoiesis, a modulator of immune responses, and a tumor suppressor in leukemia.2,3 During hematopoiesis, Ikaros plays major roles during at least 2 different stages of differentiation: (1) at the level of the lymphoid-primed multipotent progenitor (LMPP), where the absence of Ikaros results in failed lymphoid differentiation4,5 ; and (2) at early stages of T- and B-cell differentiation—in double-positive thymocytes and in pre-B cells—where reduced Ikaros function is associated with malignant transformation in both mice and humans.6-8 The importance of Ikaros for clinical hematology and oncology was underscored when Ikzf1 haploinsufficiency was shown to result in acute lymphoblastic leukemia with a high risk for relapse.9 Our knowledge of the regulatory network that controls Ikzf1 transcription is very limited. Due to the requirement for fine-tuned control of Ikaros levels for normal hematopoiesis, it is logical for its transcription to be controlled by complex mechanisms that are specific for each stage of differentiation. Dissecting the mechanisms for transcriptional regulation of Ikzf1 expression is thus of paramount importance for understanding hematopoiesis and malignant transformation and will profoundly impact clinical science.
Using a mouse transgenic reporter approach, combined with chromatin immunoprecipitation and next-generation sequencing (ChIP-SEQ), Yoshida et al1 functionally evaluate the highly conserved enhancer regions of the Ikzf1 locus. Previous studies by this group identified 10 putative regulatory regions at the Ikzf1 locus.10 The evolutionarily conserved putative Ikzf1 enhancers were tested in vivo for their ability to promote transcription of a green fluorescent protein (GFP) reporter under the control of the main Ikzf1 promoter. For each of these, a putative enhancer reporter cassette was produced and used to generate multiple transgenic mouse lines. GFP expression was evaluated in peripheral leukocytes and compared with GFP expression in a reporter cassette driven by the GFP promoter alone.
The Ikzf1 enhancers were evaluated for their ability to (1) counteract transcriptionally repressive chromatin; (2) increase transcription levels in permissive chromatin; and (3) stimulate cell type–specific transcription during different stages of lymphoid and myeloid lineage differentiation. The results identified enhancers F, H, and I as potent stimulators of transcription, but only in permissive chromatin, thus acting similarly to classical enhancers that promote initiation and elongation of transcription. In contrast, enhancers D and J were efficient in counteracting repressive chromatin, which likely is the result of recruitment of epigenetic factors that promote a transcriptionally permissive chromatin state to that region.
All of the Ikzf1 enhancers were able to stimulate the Ikzf1 promoter-based expression in B and myeloid cells. However, only 2 enhancers (D and H) were able to stimulate transcription in T cells as well. The D enhancer was the only one capable of stimulating GFP expression above the basal level in the LMPP that marks the earliest stage of lymphoid differentiation. Further analysis revealed that enhancer D is critical in counteracting repressive chromatin at the Ikzf1 locus and for maintaining a high level of transcription, and these functions could not be compensated by any other Ikzf1 enhancers. The authors further dissected the enhancer D and identified subdomains that confer stage-specific expression in T-lineage cells.
Although individual enhancers were capable of stimulating Ikzf1 expression, their activity could not replicate the activity of the wild-type endogenous Ikzf1 locus. The endogenous gene expression pattern in hematopoietic cells and in the neuronal lineage was ensured when 9 of the 10 conserved Ikzf1enhancers were combined in a miniregulatory locus.
The authors conclude these studies by analyzing previous ChIP-SEQ data to identify a network of transcription factors that bind in vivo at the Ikzf1 enhancers. These analyses revealed binding of HEB, runt-related transcription factor 1 (Runx1), T-cell factor 1, and Ikaros in thymocytes; avian myelocytomatosis oncogene (c-Myc) and avian erythroblastosis virus E26 oncogene 1 (Ets-1) in B cells; and GATA binding protein 1 (GATA1), GATA2, and stem cell leukemia/T cell acute lymphocytic leukemia 1 (SCL/Tal1) in erythroid precursors. In addition, motif search for transcription binding sites at enhancers D and H identified enrichment of binding sites for several transcriptional factors with important roles in hematopoiesis; eg, Runx, Homeobox A9 (HoxA9), special AT-rich sequence binding protein 1 (Satb1), Interferon regulatory factor-1 (Irf1), Irf4, CCAAT/enhancer binding protein–α (C/EBP-α), C/EBP-β, myocyte enhancer factor 2C (MEF2C), and E2A. The proposed working model by Yoshida et al, based on these findings, is outlined (see figure).
What are some of the implications? Next-generation sequencing has identified inactivating deletions and mutations at the Ikzf1 locus in a large subset of B-cell precursor acute lymphoblastic leukemia (B-ALL) and in early T-cell ALL in humans. These genetic alterations that result in reduced Ikzf1 activity are poor prognostic indicators in pre-B-ALL. The identification of Ikzf1 enhancers that are essential for optimal Ikzf1 expression provides an additional tool to identify potential prognostic markers. The obvious next step would include sequencing Ikzf1 enhancer elements and correlation of potential mutations and/or polymorphism in these regions with the development and/or outcome of leukemia. With the rapid development of next-generation sequencing technology and the decreased cost of sequencing, these assays are quite feasible and may yield important diagnostic information.
The identification of a network of transcription factors that positively regulates Ikzf1 expression provides an opportunity to uncover larger signaling pathways that control normal and malignant hematopoiesis. Besides the obvious impact on scientific advances in the field, this could also have important therapeutic implications. The modulation of signaling pathways that control Ikzf1 expression could be a powerful tool for the treatment of hematopoietic malignancies and some immunological disorders. This story is just developing.
Conflict-of-interest disclosure: The author declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal