Key Points
DNA methylation profile of Runx1 locus correlates with transcriptional activity and promoter usage during blood development.
Distal promoter hypomethylation is a novel signature of definitive hematopoiesis and is promoted in vitro by direct interaction with HoxB4.
Abstract
The transcription factor Runx1 (AML1) is a central regulator of hematopoiesis and is required for the formation of definitive hematopoietic stem cells (HSCs). Runx1 is alternatively expressed from two promoters: the proximal (P2) prevails during primitive hematopoiesis, while the distal (P1) dominates in definitive HSCs. Although some transcription factor binding sites and cis-regulatory elements have been identified, a mechanistic explanation for the alternative promoter usage remains elusive. We investigated DNA methylation of known Runx1 cis-elements at stages of hematopoietic development in vivo and during differentiation of murine embryonic stem cells (ESCs) in vitro. In vivo, we find loss of methylation correlated with the primitive to definitive transition at the P1 promoter. In vitro, hypomethylation, acquisition of active chromatin modifications, and increased transcriptional activity at P1 are promoted by direct interaction with HOXB4, a transcription factor that confers definitive repopulation status on primitive hematopoietic progenitors. These data demonstrate a novel role for DNA methylation in the alternative promoter usage at the Runx1 locus and identify HOXB4 as a direct activator of the P1 promoter. This epigenetic signature should serve as a novel biomarker of HSC potential in vivo, and during ESC differentiation in vitro.
Introduction
Understanding the molecular pathways governing the development of mammalian hematopoietic stem cells (HSCs) is crucial to identifying methods for their derivation from pluripotent cell sources. Of the key players involved in the formation of HSCs, the runt-related transcription factor Runx1 plays an essential role. Without Runx1, hemogenic endothelium of the aorta gonad mesonephros does not undergo hematopoietic transition, and Runx1 knockout embryos fail to survive past 12.5 days gestation with a complete loss of definitive hematopoiesis.1-4 This defect is mirrored during hematopoietic differentiation of embryonic stem cells (ESCs) in vitro, underscoring the role of Runx1 as a fundamental regulator of the hematopoietic program during development.5,6
Runx1 serves as the α subunit of the core-binding factor complex and is the most common translocation in acute myeloid leukemia in humans.7,8 Defining regulation of the Runx1 locus is necessary for understanding the complex functional roles and tightly regulated activity of Runx1 during HSC development and hematopoietic malignancy. Runx1 transcription is controlled by two developmentally regulated alternative promoters and an intronic enhancer (+23) element.9-12 Intriguingly, promoter usage follows a pattern whereby the proximal (P2) promoter initiates early in primitive hematopoiesis, while the distal (P1) promoter–driven transcription appears later in definitive hematopoietic cells.13,14 Strikingly, distal promoter predominance is temporally associated with peak HSC expansion in the fetal liver (FL) and continues into adult marrow where purified HSCs use the distal promoter almost exclusively.12,14-16 While Runx1 core promoter elements fail to restrict reporter expression to the hematopoietic lineage, the intronic +23 enhancer is sufficient to drive hematopoietic specificity via interaction with Gata2, Ets factors, and the SCL/Lmo2/Ldb1 complex.9,10 Although much has been learned about Runx1 promoter and enhancer usage, the precise mechanisms responsible for the observed promoter switch are not understood.
DNA methylation is an epigenetic regulatory mechanism that contributes prominently to embryonic development and lineage commitment in the hematopoietic system.17-19 Recent advances in genome-wide analysis of methylation have identified tissue-specific differentially methylated regions (TDMRs) as dynamic regulators of gene expression during development and disease.20 Further evidence suggests that intronic enhancers are frequently TDMRs involved in lineage commitment and can influence the usage of alternative promoters.20,21 In spite of these observations, methylation of the Runx1 regulatory elements has not yet been assessed in the context of hematopoietic development, and relatively little is known about the mechanisms underlying the Runx1 promoter switch.
Here, we examine the DNA methylation status of Runx1 regulatory elements at stages of hematopoietic development in vivo and during hematopoietic differentiation of ESCs in vitro. We find that the P2 promoter is unmethylated in all cell types examined, including pluripotent murine embryonic stem cells (mESCs). In accordance with its role as a hematopoietic-specific enhancer, a striking loss of methylation is observed at the +23 enhancer element upon commitment to the hematopoietic lineage in vivo and in vitro. We show that hypomethylation of the distal promoter region is correlated with definitive HSC potential in vivo and can be promoted during in vitro hematopoietic differentiation by HOXB4. Concordantly, HOXB4 overexpression promotes P1 transcription and an increased P1/P2 messenger RNA (mRNA) ratio. Chromatin immunoprecipitation (chIP) analyses in mESC-derived hematopoietic cells overexpressing HOXB4 show that P1 is preferentially bound by HOXB4, acquires histone modifications permissive to transcription, and undergoes a decrease in occupancy by the maintenance methyltransferase DNMT1. Thus, decreased methylation, acquisition of active chromatin modifications, and increased transcriptional activity at the P1 promoter is promoted in vitro via physical interaction with HOXB4.
Materials and methods
Information concerning cell culture, ESC differentiation, retroviral transduction, embryo dissection, cell isolation and fractionation, bisulfite sequencing, methylation analysis, quantitative reverse transcriptase polymerase chain reaction, and chIP is provided in the supplemental Data.
Statistics
Statistical analysis of methylation between individual populations was performed by using the online quantification tool for methylation analysis program (http://quma.cdb.riken.jp/). Differences between grouped methylation data were determined by using an unpaired 2-tailed Student t test, with P values less than .05 considered significant. All other analyses were performed by using a Student t test, with P values less than .05 considered significant.
Results
Proximal Runx1 P2 promoter is unmethylated regardless of lineage or developmental status
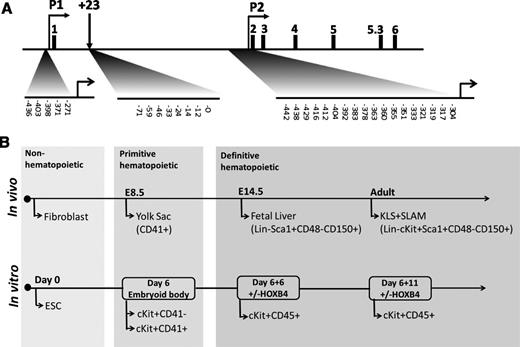
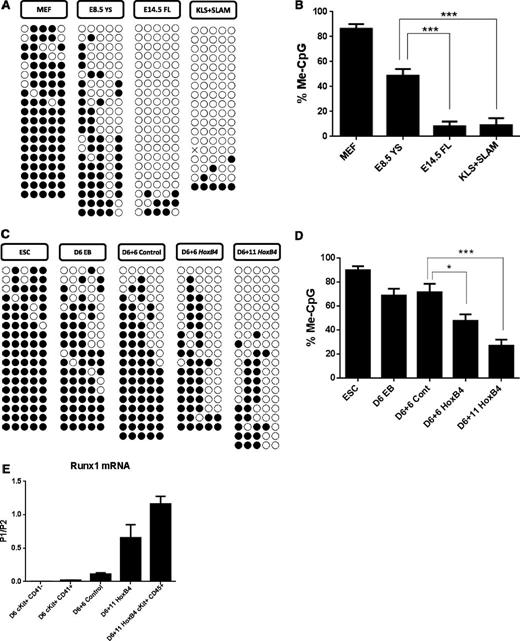
The exceptionally large size of the Runx1 locus (224 kb) limited our ability to perform locus-wide exploratory bisulfite sequencing. We thus focused on Runx1 regulatory elements previously confirmed to exhibit hematopoietic activity10 (Figure 1A; supplemental Table 1). The Runx1 P2 promoter is the most CpG dense (63% GC, 1.03 CpG observed [Obs]/global expected [Exp]) of the three Runx1 elements analyzed and the only one to contain a classic CpG island (GC >50%, Obs/Exp P > .6). To determine whether the proximal Runx1 promoter is differentially methylated during hematopoietic development in vivo, we analyzed bisulfite-treated genomic DNA from E14.5 mouse embryonic fibroblast (MEF) cells, E8.5 yolk sac (YS) CD41+, E14.5 FL Lin-Sca-1+CD48−CD150+, and adult marrow Lin-c-Kit+Sca-1+CD150+CD48− (KLS + SLAM) cells representing nonhematopoietic, primitive hematopoietic, and two stages of definitive HSCs, respectively (Figure 1B; supplemental Figure 1A-B). To parallel this analysis in vitro using mESCs, we used an established 2-stage differentiation process (Figure 1B). mESCs were first differentiated as embryoid bodies (EBs) for 6 days to obtain primitive c-Kit+CD41+ progenitors (supplemental Figure 2A). Since HOXB4 overexpression is an efficient method of obtaining ESC-derived hematopoietic progenitors capable of robust long-term reconstitution posttransplant,22 we used overexpression of HOXB4 and coculture on hematopoietic supportive OP9 stromal cells to obtain mESC-derived definitive HSCs. Day 6 EB cells were infected with MSCV-HOXB4-IRES-GFP or MSCV-IRES-GFP control virus and cocultured on OP9 cells. The GFP+c-Kit+CD45+ fraction was isolated from HOXB4 and control cocultures after 6 days (6 + 6 days) and from HOXB4 cocultures at 11 days (6 + 11 days) at which point no hematopoietic cells were observed in the control cocultures (supplemental Figure 2B).
Runx1 genomic locus and cell populations for bisulfite analysis. (A) Murine Runx1 locus and CpGs analyzed by bisulfite sequencing. For P1 and P2, CpG number represents distance from transcription start site. For +23, CpG number represents distance from the 3′ end of the polymerase chain reaction (PCR) amplicon. (B) Cell populations isolated in vivo and from ESC differentiation strategy for bisulfite analysis.
Runx1 genomic locus and cell populations for bisulfite analysis. (A) Murine Runx1 locus and CpGs analyzed by bisulfite sequencing. For P1 and P2, CpG number represents distance from transcription start site. For +23, CpG number represents distance from the 3′ end of the polymerase chain reaction (PCR) amplicon. (B) Cell populations isolated in vivo and from ESC differentiation strategy for bisulfite analysis.
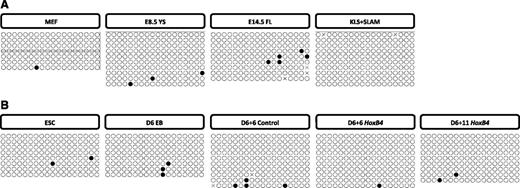
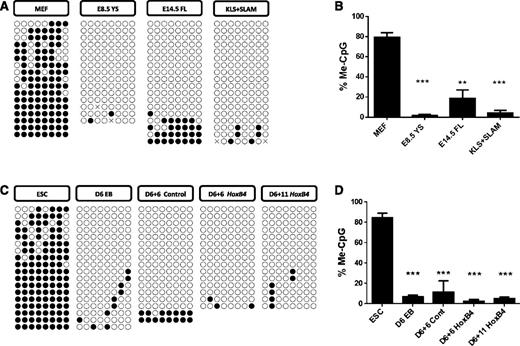
In vivo, we find the P2 promoter to be unmethylated in MEFs, a nonhematopoietic control (0.8% ± 0.79%) (Figure 2A). No change in P2 promoter methylation was associated with primitive hematopoiesis because P2 was also unmethylated in E8.5 YS CD41+ cells (1.7% ± 0.85%) (Figure 2A). This absence of P2 promoter methylation continues in E14.5 FL Lin-Sca-1+CD48−CD150+ cells (3.1% ± 1.63%), and in adult marrow KLS + SLAM (0% ± 0%) (Figure 2A), indicating that the Runx1 P2 promoter does not exhibit differential methylation between lineages or during hematopoietic development in vivo. Although previous reports indicate a P1 bias in FL and adult HSCs,12,15,16 our data indicate that the methylation status of the P2 promoter does not establish this bias.
Bisulfite analysis of proximal Runx1 promoter. (A) Methylation patterns of the Runx1 P2 promoter in cells derived from E14.5 MEFs, E8.5 YS CD41+ (E8.5 YS), E14.5 FL Lin-Sca-1+CD48−CD150+ (E14.5 FL), and adult marrow Lin-c-Kit+Sca-1+CD150+CD48− (KLS + SLAM). Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and solid circles represent methylated CpGs. (B) Methylation patterns in mESCs, day 6 EB c-Kit+CD41+ (D6 EB), and from OP9 cocultures: GFP+c-Kit+CD45+ cells isolated from IRES-GFP control group at day 6 (D6 + 6 control) and HOXB4-IRES-GFP group at day 6 (D6 + 6 HoxB4) and day 11 (D6 + 11 HoxB4).
Bisulfite analysis of proximal Runx1 promoter. (A) Methylation patterns of the Runx1 P2 promoter in cells derived from E14.5 MEFs, E8.5 YS CD41+ (E8.5 YS), E14.5 FL Lin-Sca-1+CD48−CD150+ (E14.5 FL), and adult marrow Lin-c-Kit+Sca-1+CD150+CD48− (KLS + SLAM). Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and solid circles represent methylated CpGs. (B) Methylation patterns in mESCs, day 6 EB c-Kit+CD41+ (D6 EB), and from OP9 cocultures: GFP+c-Kit+CD45+ cells isolated from IRES-GFP control group at day 6 (D6 + 6 control) and HOXB4-IRES-GFP group at day 6 (D6 + 6 HoxB4) and day 11 (D6 + 11 HoxB4).
The absence of P2 promoter methylation was mirrored in vitro in undifferentiated mESCs (1.4% ± 0.91%) and did not change in day 6 EB c-Kit+CD41+ cells (2.1% ± 1.02%) (Figure 2B). P2 remained unmethylated in day 6 + 6 OP9 cocultures in both control c-Kit+CD45+ cells (2.2% ± 1.79%) and HOXB4 overexpressing cells at day 6 + 6 (0.6% ± 0.56%) and day 6 + 11 (1.2% ± 0.82%) (Figure 2B). These observations are consistent with previous reports that CpG-dense core promoter regions are predominantly unmethylated,23,24 and that P2 expression exhibits a lower degree of lineage-restricted expression than P1 during development.25 Indeed, global DNA methylation data released by the ENCODE Consortium confirms a foci of hypomethylation at the P2 promoter in multiple cell lineages.26
Hypomethylation of the +23 enhancer correlates with the hematopoietic lineage
The recently described intronic +23 enhancer element has been shown to drive hematopoietic-specific transcription.10 Sequence analysis of this region shows low CpG density and does not specify a classic CpG island (58% GC, 0.31 Obs/Exp). In vivo, we find the +23 enhancer is highly methylated in MEFs (79.4% ± 4.66%), but methylation is significantly decreased in E8.5 YS CD41+ cells (1.7% ± 1.22%), E14.5 FL Lin-Sca-1+CD48−CD150+ (18.8% ± 8.29%), and adult KLS + SLAM (4.2% ± 2.54%) (Figure 3A-B). These results demonstrate that +23 enhancer hypomethylation occurs during embryonic hematopoiesis and continues into fetal and adult hematopoiesis.
Bisulfite analysis of +23 enhancer methylation. (A) Methylation patterns of the Runx1 +23 enhancer in cells from in vivo–derived E14.5 MEFs, E8.5 YS CD41+ (E8.5 YS), E14.5 FL Lin-Sca-1+CD48−CD150+ (E14.5 FL), and adult marrow Lin-c-Kit+Sca-1+CD150+CD48− (KLS + SLAM). Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and solid circles represent methylated CpGs. (B) Quantification of percent CpG methylation at +23 in hematopoietic populations derived in vivo. ***P < .001; **P < .01. (C) Methylation patterns in mESC day 6 EB c-Kit+CD41+ (D6 EB) and from OP9 cocultures: GFP+c-Kit+CD45+ cells isolated from IRES-GFP control group at day 6 (D6 + 6 control) and HOXB4-IRES-GFP group at day 6 (D6 + 6 HoxB4) and day 11 (D6 + 11 HoxB4). (D) Quantification of percent CpG methylation at +23 in cell populations isolated during hematopoietic differentiation of mESCs. ***P < .001.
Bisulfite analysis of +23 enhancer methylation. (A) Methylation patterns of the Runx1 +23 enhancer in cells from in vivo–derived E14.5 MEFs, E8.5 YS CD41+ (E8.5 YS), E14.5 FL Lin-Sca-1+CD48−CD150+ (E14.5 FL), and adult marrow Lin-c-Kit+Sca-1+CD150+CD48− (KLS + SLAM). Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and solid circles represent methylated CpGs. (B) Quantification of percent CpG methylation at +23 in hematopoietic populations derived in vivo. ***P < .001; **P < .01. (C) Methylation patterns in mESC day 6 EB c-Kit+CD41+ (D6 EB) and from OP9 cocultures: GFP+c-Kit+CD45+ cells isolated from IRES-GFP control group at day 6 (D6 + 6 control) and HOXB4-IRES-GFP group at day 6 (D6 + 6 HoxB4) and day 11 (D6 + 11 HoxB4). (D) Quantification of percent CpG methylation at +23 in cell populations isolated during hematopoietic differentiation of mESCs. ***P < .001.
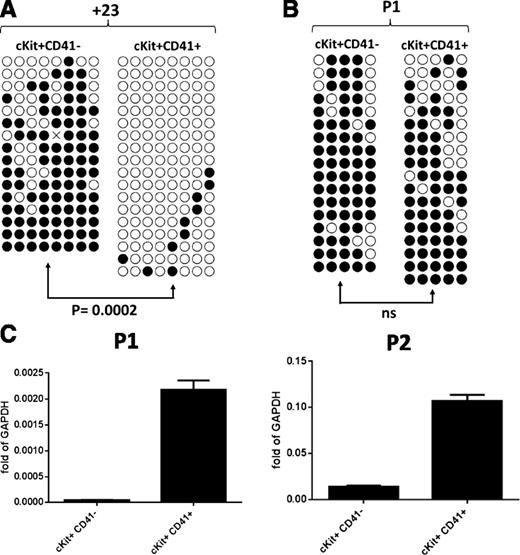
This correlation of +23 hypomethylation and the hematopoietic lineage also occur during hematopoietic differentiation of mESCs. The +23 enhancer is methylated to a high degree in undifferentiated mESCs (84.7% ± 4.23%), followed by a significant decrease in day 6 EB c-Kit+CD41+ hematopoietic cells (6.9% ± 1.81%) that is maintained in OP9 cocultured control c-Kit+CD41+ cells at day 6 + 6 (11.4% ± 11.1%) and in HOXB4 overexpressing cells at both day 6 + 6 (2.5% ± 1.81%) and day 6 +11 (5% ± 1.64%) (Figure 3C-D). To further define the hematopoietic specificity of +23 hypomethylation, we compared the +23 methylation profile of day 6 EB c-Kit+CD41− cells to that of day 6 c-Kit+CD41+ cells, since CD41 is the earliest known marker of hematopoiesis.27,28 We found that +23 hypomethylation strongly correlated with the acquisition of hematopoietic fate as defined by CD41 (70.9% ± 6.1% vs 6.9% ± 1.81%; P = .0002) (Figure 4A). In contrast, there is no statistically significant difference in the methylation of the distal promoter between these populations (Figure 4B).
Bisulfite analysis and Runx1 expression in day 6 EB subpopulations. (A) Methylation patterns of the Runx1 +23 enhancer in c-Kit+CD41− and c-Kit+CD41+ cells from day 6 EBs. Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and filled circles represent methylated CpGs. (B) Methylation patterns of the Runx1 P1 promoter in c-Kit+CD41− and c-Kit+CD41+ cells from day 6 EBs. (C) Quantitative reverse transcriptase polymerase chain reaction analysis of Runx1 P1 and P2 mRNA isoforms in c-Kit+CD41− and c-Kit+CD41+ cells from day 6 EBs. ns, not significant.
Bisulfite analysis and Runx1 expression in day 6 EB subpopulations. (A) Methylation patterns of the Runx1 +23 enhancer in c-Kit+CD41− and c-Kit+CD41+ cells from day 6 EBs. Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and filled circles represent methylated CpGs. (B) Methylation patterns of the Runx1 P1 promoter in c-Kit+CD41− and c-Kit+CD41+ cells from day 6 EBs. (C) Quantitative reverse transcriptase polymerase chain reaction analysis of Runx1 P1 and P2 mRNA isoforms in c-Kit+CD41− and c-Kit+CD41+ cells from day 6 EBs. ns, not significant.
Enhancer methylation has been shown to influence gene expression.29 We examined the expression of the P1 and P2 mRNA in day 6 EB c-Kit+CD41− and c-Kit+CD41+ cells and found that both P1 and P2 transcriptional activities are increased in c-Kit+CD41+ cells (Figure 4C). Thus, +23 methylation influences the activity of both Runx1 promoters in a manner that correlates with CD41 acquisition.
Distal Runx1 P1 promoter is hypomethylated in HSCs in vivo
Several studies have observed that expression of the P1 Runx1 mRNA isoform is specifically upregulated in definitive hematopoietic cells during development and comprises the majority of Runx1 transcript in HSCs.12,15,16 Although the P2 promoter is CpG dense and structurally similar to housekeeping promoter elements,9 the P1 promoter is more complex in terms of transcription factor binding sites and comparatively CpG poor (46% GC, 0.25 Obs/Exp). Consistent with the definitive hematopoiesis-specific activity of the P1 promoter, a high degree of methylation is observed in MEFs (86.3% ± 3.76%) (Figure 5A-B). Compared with MEFs, there is a modest yet statistically significant decrease in P1 promoter methylation in E8.5 YS CD41+ cells (48.6% ± 5.45) (Figure 5A-B). However, an even greater degree of P1 hypomethylation is observed in definitive HSCs, where E14.5 FL Lin-Sca-1+CD48−CD150+ (8.1% ± 3.89%) and KLS + SLAM (9% ± 5.65%) cells exhibit a significantly lower level of P1 methylation compared with E8.5 YS CD41+ cells (Figure 5A-B).
Bisulfite analysis and mRNA expression from Runx1 P1 promoter. (A) Methylation patterns of the Runx1 P1 promoter in cells from in vivo–derived E14.5 MEFs, E8.5 YS CD41+ (E8.5 YS), E14.5 FL Lin-Sca-1+CD48−CD150+ (E14.5 FL), and adult marrow Lin-c-Kit+Sca-1+CD150+CD48− (KLS + SLAM). Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and solid circles represent methylated CpGs. (B) Quantification of percent CpG methylation at P1 in hematopoietic populations derived in vivo. ***P < .001. (C) Methylation patterns in mESCs, day 6 EB c-Kit+CD41+ (D6 EB) and from OP9 cocultures: GFP+c-Kit+CD45+ cells isolated from IRES-GFP control group at day 6 (D6 + 6 control) and HOXB4-IRES-GFP group at day 6 (D6 + 6 HoxB4) and day 11 (D6 + 11 HoxB4). (D) Quantification of percent CpG methylation at P1 in cell populations isolated during hematopoietic differentiation of mESCs. ***P < .001; *P < .05. (E) P1/P2 mRNA levels after normalization to Gapdh over the course of hematopoietic differentiation in vitro.
Bisulfite analysis and mRNA expression from Runx1 P1 promoter. (A) Methylation patterns of the Runx1 P1 promoter in cells from in vivo–derived E14.5 MEFs, E8.5 YS CD41+ (E8.5 YS), E14.5 FL Lin-Sca-1+CD48−CD150+ (E14.5 FL), and adult marrow Lin-c-Kit+Sca-1+CD150+CD48− (KLS + SLAM). Sequencing reactions of individual amplicons are represented by each row of circles. Open circles denote unmethylated CpGs, and solid circles represent methylated CpGs. (B) Quantification of percent CpG methylation at P1 in hematopoietic populations derived in vivo. ***P < .001. (C) Methylation patterns in mESCs, day 6 EB c-Kit+CD41+ (D6 EB) and from OP9 cocultures: GFP+c-Kit+CD45+ cells isolated from IRES-GFP control group at day 6 (D6 + 6 control) and HOXB4-IRES-GFP group at day 6 (D6 + 6 HoxB4) and day 11 (D6 + 11 HoxB4). (D) Quantification of percent CpG methylation at P1 in cell populations isolated during hematopoietic differentiation of mESCs. ***P < .001; *P < .05. (E) P1/P2 mRNA levels after normalization to Gapdh over the course of hematopoietic differentiation in vitro.
HoxB4 alters P1 promoter methylation and transcriptional activity in vitro
We next determined whether this in vivo epigenetic signature of definitive HSCs is replicated during mESC differentiation. Like the +23 enhancer, P1 is highly methylated in undifferentiated mESCs (90% ± 3.3%) (Figure 5C-D). Paralleling our results in vivo, a modest decrease is observed in day 6 EB c-Kit+CD41+ cells (68.9% ± 5.65%) and in OP9 cocultured control c-Kit+CD45+ cells at day 6 + 6 (71.6% ± 7.06%) (Figure 5C-D). However, in OP9 cocultured c-Kit+CD45+ cells overexpressing HOXB4, we observe a significant decrease in P1 methylation compared with control cells at day 6 + 6 (47.8% ± 5.4%), followed by a further decrease by day 6 + 11 (27% ± 5.08%), corresponding to a population shown to possess more robust hematopoietic repopulating potential after adoptive transfer in vivo (Figure 5C-D). These data demonstrate that the Runx1 P1 promoter is methylated in pluripotent mESCs and remains methylated in the first wave of c-Kit+CD41+ hematopoietic progenitors. Maturation to c-Kit+CD45+ progenitors on OP9 alone does not change this methylation profile, whereas overexpression of HOXB4 during this process results in decreased P1 methylation.
To determine whether P1 hypomethylation is linked to promoter switching during differentiation, we examined relative P1 vs P2 mRNA levels using isoform-specific quantitative reverse transcriptase polymerase chain reaction. As expected, P2 dominates in primitive EB-derived cell populations and in OP9 control cocultures (Figure 5E). However, the P1:P2 ratio is higher in HOXB4-overexpressing OP9 cocultures, and even higher in purified c-Kit+CD45+ hematopoietic cells overexpressing HOXB4 (Figure 5E). These data suggest that HOXB4-induced P1 hypomethylation is associated with a consequent increase in the P1:P2 mRNA ratio in vitro.
HoxB4 directly activates the Runx1 P1 promoter
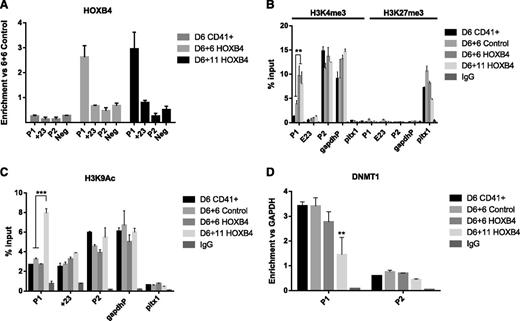
It was unclear whether the mechanism underlying HOXB4-mediated activation of P1 was direct or indirect. To answer this question, we performed chIP analysis in ESC-derived hematopoietic cells during differentiation to examine the level of HOXB4 binding at Runx1 regulatory regions. We find that HOXB4 preferentially binds the P1 promoter upon overexpression at day 6 + 6 and day 6 + 11, thus identifying a direct role for HOXB4 in the modulation of P1 transcriptional activity (Figure 6A).
Runx1 chIP analysis. (A) Quantitative PCR (qPCR) analysis of chIP performed against HOXB4. Data were normalized to the percent of pre-IP input for each sample and are expressed as the fold change vs the day 6 + 6 (D6 + 6) control population. Data are representative of at least two independent IPs. (B) qPCR analysis of chIP performed against H3K4me3 and H3K27me3. Data are expressed as the percent of pre-IP input for each sample and are representative of at least 2 independent IPs. **P < .01. (C) qPCR analysis of chIP performed against H3K9Ac. Data are expressed as the percent of pre-IP input for each sample and are representative of at least 2 independent IPs. ***P < .001. (D) qPCR analysis of chIP performed against Dnmt1. Data are expressed as the fold change vs Gapdh control locus for each sample and are representative of at least 2 independent IPs. **P < .01.
Runx1 chIP analysis. (A) Quantitative PCR (qPCR) analysis of chIP performed against HOXB4. Data were normalized to the percent of pre-IP input for each sample and are expressed as the fold change vs the day 6 + 6 (D6 + 6) control population. Data are representative of at least two independent IPs. (B) qPCR analysis of chIP performed against H3K4me3 and H3K27me3. Data are expressed as the percent of pre-IP input for each sample and are representative of at least 2 independent IPs. **P < .01. (C) qPCR analysis of chIP performed against H3K9Ac. Data are expressed as the percent of pre-IP input for each sample and are representative of at least 2 independent IPs. ***P < .001. (D) qPCR analysis of chIP performed against Dnmt1. Data are expressed as the fold change vs Gapdh control locus for each sample and are representative of at least 2 independent IPs. **P < .01.
We next explored changes in chromatin organization over the time course of hematopoietic differentiation. Bivalency of lineage-specific genes is a hallmark of pluripotency and involves the co-localization of active (H3K4me3) and repressive (H3K27me3) histone modifications, which subsequently resolve to the presence of one or the other during lineage commitment.30 We find that H3K27me3 is uniformly absent at P1, +23, and P2 as compared with a silenced control region (Pitx1) at all time points, demonstrating that the Runx1 locus is primed for activation early in the hematopoietic lineage and is not altered by HOXB4 overexpression (Figure 6B). Consistent with our expression data, H3K4me3 is enriched at the P2 promoter and an active control locus (Gapdh) over the course of differentiation and is not altered by HOXB4 (Figure 6B). Conversely, H3K4me3 presence at the P1 promoter is significantly increased upon overexpression of HOXB4 by day 6 + 6 and is maintained through day 6 + 11 (Figure 6B). Finally, we find that H3K9Ac—a mark of actively transcribed promoters—is present throughout differentiation at P2 but is significantly increased at P1 only in day 6 + 11 HoxB4 overexpressing cells (Figure 6C), a finding that is consistent with both our methylation and expression data in that this time point coincides with the highest degree of P1 hypomethylation and maximal level of P1 expression in vitro.31 From these collective results, we conclude that HOXB4 preferentially binds to the Runx1 P1 promoter and stimulates transcription via decreased methylation and establishment of a permissive chromatin state in ESC-derived hematopoietic cells.
DNMT1 occupancy at P1 is decreased in HOXB4 overexpressing cells
DNA methylation patterns are either maintained during replication via the activity of the maintenance methyltransferase DNMT1, or established de novo by DNMT3a and DNMT3b.32-34 To determine whether the loss of methylation at P1 is accompanied by decreased interaction with members of the DNA methyltransferase family, we measured the level of DNMT1, DNMT3a, and DNMT3b occupancy at the Runx1 promoters. We did not detect binding of DNMT3a or DNMT3b to P1 (data not shown); however, we found that DNMT1 did interact with P1, and this interaction was significantly decreased at day 6 + 11 in HOXB4-overexpressing cells (Figure 6D). As expected, occupancy was universally low at the hypomethylated P2 promoter at all time points (Figure 6D). These data are consistent with a mechanism in which DNMT1 is occluded from accessing the P1 promoter in the presence of HOXB4, resulting in a gradual loss of established methylation patterns over subsequent cell divisions.
Runx1 P1 methylation as signature of definitive HSCs
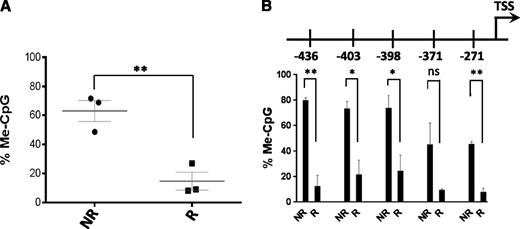
Finally, to compare our in vivo and in vitro observations that P1 methylation is correlated with definitive HSCs during development, we grouped all cell populations studied on the basis of whether they are known to possess hematopoietic repopulating capacity, and we compared mean levels of P1 promoter methylation. This comparison indicated that P1 promoter hypomethylation correlated with repopulating capacity in a similar fashion in vivo and in vitro (Figure 7A). To increase the resolution of P1 promoter demethylation, we applied the same grouped comparison with the individual CpG dinucleotides in the P1 promoter and found that overall methylation was significantly decreased for each CpG, with the exception of the CpG located at position −371 relative to the P1 transcription start site. In particular, the CpGs located at positions −436 and −271 were the most significantly different between repopulating and non-repopulating cell types (Figure 7B).
Comparison of P1 methylation in repopulating and non-repopulating cell types. (A) Percent CpG methylation at P1 promoter in repopulating (R) and non-repopulating (NR) cell populations. Bar indicates the mean. **P < .01 (unpaired Student t test). (B) Percent methylation at individual CpGs within P1 promoter in repopulating and non-repopulating cell populations. Bar indicates the mean. **P < .01; *P < .05 (unpaired Student t test). ns, not significant.
Comparison of P1 methylation in repopulating and non-repopulating cell types. (A) Percent CpG methylation at P1 promoter in repopulating (R) and non-repopulating (NR) cell populations. Bar indicates the mean. **P < .01 (unpaired Student t test). (B) Percent methylation at individual CpGs within P1 promoter in repopulating and non-repopulating cell populations. Bar indicates the mean. **P < .01; *P < .05 (unpaired Student t test). ns, not significant.
Discussion
Here, we identify previously undescribed changes in DNA methylation at Runx1 regulatory regions during hematopoietic development. These changes are associated with commitment to the hematopoietic lineage, and they distinguish definitive repopulating HSC populations from earlier, primitive non-repopulating cells. We find that the CpG-dense P2 promoter is unmethylated in mESCs and remains unmethylated regardless of lineage and stage of hematopoietic development. Conversely, the +23 intronic enhancer is methylated in mESCs, nonhematopoietic fibroblasts, and c-Kit+CD41− cells from day 6 EBs but is dramatically hypomethylated upon acquisition of CD41 and remains unmethylated throughout hematopoietic development. Importantly, we demonstrate that hypomethylation of the P1 Runx1 promoter is specific to cell populations enriched in definitive repopulating capacity in vivo. Moreover, in mES-derived c-Kit+CD41+ hematopoietic progenitors, the P1 promoter remains methylated at levels similar to those observed in E8.5 YS. Overexpression of HOXB4 results in a significant decrease in P1 methylation consistent with its ability to generate mESC-derived hematopoietic progenitors capable of long-term repopulation in transplant recipients.22 Thus, our results identify hypomethylation of the distal Runx1 promoter as a novel epigenetic signature of repopulating hematopoietic cells during development and provide critical insight into the dynamic epigenetic changes influencing the Runx1 locus.
The developmental processes leading to the formation of definitive HSCs rely on the properly orchestrated activity of a complex network of critical transcription factors to guide genetic programs associated with differentiation. Runx1 is a critical transcription factor involved in the development of definitive HSCs1,2,5 and, not surprisingly, is frequently disregulated in hematopoietic malignancy.7,8 A diverse array of Runx1 mRNA isoforms have been identified35 and arise through a combination of alternative splicing as well as alternate promoter usage.9,12,13,36 As methods for genome-wide analysis of CpG methylation have improved, a growing number of TDMRs associated with normal and abnormal development have been identified.37 Previously, it was unknown whether the Runx1 regulatory regions were TDMRs. In the case of the Runx1 P2 promoter, our data now indicate it is not a TDMR because P2 is unmethylated in a wide array of cell types. This observation is consistent with previous reports of other CpG-rich core promoters23,24 and is supported by genome-wide DNA methylation profiles released by the ENCODE Consortium.26 That the P2 methylation pattern is established at the pluripotent stage and does not change in differentiated cell types or during hematopoietic development suggests that methylation of P2 does not influence lineage-specific changes in P2 transcription during development. It seems unlikely that P2 methylation acts as an on/off switch, since the P2 isoform is detected in a diverse array of cell types,25 including undifferentiated mESCs,38 and the P2 promoter remains unmethylated in FL and adult HSCs, even though P1 is the dominant promoter in these populations.12,15,16 Therefore, our observations suggest that P2 methylation is not involved in the lineage restriction of P2 during development. Intriguingly, intragenic TDMR downstream of P2 have been identified, raising the possibility that methylation at these regions could have a role in the regulation of P2 activity.17
In contrast to the proximal promoter, the +23 intronic enhancer is a TDMR, and while the hematopoietic-specific activity of the Runx1 +23 enhancer element is documented,10 our data are the first to identify changes in CpG methylation at the +23 enhancer element during hematopoietic development. Previous data indicating that methylation of intronic enhancer elements influences tissue-specific gene expression29 further supports a role for +23 enhancer methylation in the transcriptional activity of Runx1. We confirm this hypothesis by clearly demonstrating that +23 hypomethylation strongly correlates with increased transcription at both Runx1 promoters at the earliest stage of hematopoietic development, although P2 remains the dominant mRNA isoform in these populations. There is evidence that intragenic DNA methylation influences alternate promoter usage21 ; however, our observations indicate that +23 hypomethylation is not a significant factor in the observed P1/P2 switch but rather acts as an epigenetic rheostat for the Runx1 locus, mediating hematopoietic-specific amplification of Runx1 expression. We cannot rule out the possibility that +23 hypomethylation facilitates a more permissive state for P1 transcription by allowing improved mRNA elongation,39 although this does not fit our observation that +23 enhancer methylation is lost early in development when P2 still dominates compared with adult HSCs.12,15 Considering our data that P1 hypomethylation results in a shift to P1-biased expression, a more probable explanation is that +23 methylation is a nonbiased regulator of transcription from both promoters, and the usage bias is determined by the epigenetic status of P1. Studies using clonal cell populations with well-defined mRNA isoform expression profiles or episomal reporters in which methylation can be artificially manipulated might help to refine the role of methylation in regulating promoter usage. However, the utility of these assays is limited by the lack of developmental and genomic context. Regardless, our data clearly demonstrate that +23 methylation influences its enhancer capacity and provides a novel epigenetic signature of the hematopoietic lineage, which can be applied to optimize methods for direct conversion of other lineages to a hematopoietic fate.40
Derivation of robust numbers of in vivo long-term repopulating HSCs from pluripotent cell sources such as ES and induced pluripotent stem cells is of therapeutic interest. Critical to the success of these efforts is the identification of signatures associated with the formation of definitive HSCs during development. Although their precise role in the regulation of gene transcription is not fully understood, TDMRs and differential DNA methylation have nonetheless proven useful for identifying differences in cell populations. This is highlighted by the use of DNA methylation patterns to determine whether somatic cells have been successfully reprogrammed to a pluripotent state.41,42 We have identified the Runx1 P1 promoter as a novel TDMR and have demonstrated that hypomethylation of this region differentiates primitive non-repopulating progenitors and definitive repopulating HSCs during embryonic development in vivo. Our observation that unmodified hematopoietic progenitors derived from mESCs do not undergo this decrease in P1 methylation suggests a failure in the epigenetic transition to adult-type definitive hematopoiesis. By overexpressing HOXB4 in ESC-derived hematopoietic cells, we show that it is possible to promote epigenetic remodeling of the Runx1 locus during differentiation of ESCs to hematopoietic cells in vitro. The hypomethylation of P1 induced by HOXB4 results in an increased P1:P2 mRNA ratio and thus links P1 hypomethylation to the P2-to-P1 promoter switch observed during hematopoietic development. Mechanistically, we show that when overexpressed, HOXB4 preferentially binds P1, supporting a model in which the epigenetic remodeling and increased transcription of P1 is mediated via physical interaction of HOXB4 with this locus. Recent HOXB4 chIP-Seq data support our findings that HOXB4 interacts with the Runx1 distal promoter, albeit using a slightly different ESC differentiation method, suggesting that this is a robust biological phenomenon.43 Whether demethylation of P1 in the presence of HoxB4 is an active or passive process remains unclear. However, our observation that P1 methylation is only slightly decreased at day 6 + 6 and becomes more pronounced by day 6 + 11 is congruent with a passive loss of methylation. This is supported by reports finding that active demethylation often occurs rapidly—within minutes or hours—and results in nearly complete demethylation of the region in question.44,45 Since HOXB4 physically binds Runx1 P1, it is conceivable that HOXB4 or a HOXB4-associated complex could physically or functionally occlude the activity of factors involved in maintaining DNA methylation, such as Dnmt1. Indeed, our data demonstrating a decrease in Dnmt1 occupancy of P1 in HOXB4-overexpressing cells are consistent with this model. Over successive rounds of DNA replication, this would result in a loss of Runx1 P1 methylation. Previous reports that differentiation-induced hypomethylation of lineage-specific CpG-poor promoters correlates with transcription-factor binding further supports this explanation.46
The fact that epigenetic remodeling of P1 could be achieved by overexpressing HOXB4 demonstrates that this in vivo epigenetic signature is valid during ESC differentiation and can potentially be replicated if the appropriate extracellular cues are applied during the differentiation process, whether in vivo or in vitro, via the induction of appropriate transcription factor circuits. Our single CpG group analysis identifies the CpGs located at −436 and −271 from the P1 transcription start site as the most significantly hypomethylated in definitive repopulating cell populations and presents an attractive target for high-throughput analysis of changes in P1 methylation during hematopoietic differentiation of pluripotent cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Troy Lund, Mark Osborn, and Istvan Szatmari for valuable advice and technical assistance.
This work was supported in part by the National Institutes of Health (NIH) Research Program Grant, National Cancer Institute P01 CA065493 and NIH Research Project Grant Program, National Institute of Allergy and Infectious Diseases R01 AI081918 (to B.R.B.); NIH Research Project Cooperative Agreement, National Heart, Lung and Blood Institute U01 HL 100407 (to M.K.); 5T32HD060536-2 awarded to the University of Minnesota Stem Cell Institute (to B.R.W.); and a grant awarded to the Children's Cancer Research Fund.
Authorship
Contribution: B.R.W. designed and performed experiments, analyzed data, and wrote the manuscript; M.I. and S.H.C. assisted in designing and performing experiments; and J.T., M.K., and B.R.B. discussed experiments and results and edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Corresponding author: Bruce R. Blazar, Department of Pediatrics, MMC 109, University of Minnesota, Minneapolis, MN 55455; e-mail: blaza001@umn.edu.
References
Author notes
J.T., M.K., and B.R.B. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal