Abstract
Acute graft-versus-host disease (GVHD) afflicts as much as 80% of all patients who receive an unrelated donor hematopoietic cell transplant (HCT) for the treatment of blood disorders, even with optimal donor HLA matching and use of prophylactic immunosuppressive agents. Of patients who develop acute GVHD, many are at risk for chronic GVHD and bear the burden of considerable morbidity and lowered quality of life years after transplantation. The immunogenetic basis of GVHD has been the subject of intensive investigation, with the classic HLA genetic loci being the best-characterized determinants. Recent information on the major histocompatibility complex (MHC) region of chromosome 6 as an important source of untyped genetic variation has shed light on novel GVHD determinants. These data open new paradigms for understanding the genetic basis of GVHD.
Introduction
The success of hematopoietic cell transplant (HCT) is influenced by many factors including HLA mismatching, major histocompatibility complex (MHC) region variation, minor histocompatibility targets of allorecognition, regulatory elements that affect gene expression, and genetic variation that affects immune responses.1-18 The individual contribution from each of these factors and pathways may be challenging to quantify in individual patients; however sophisticated tools and analysis methods are available for assessing the global impact of genetic variation on transplant outcome.
The best studied region of the genome is the MHC on chromosome 6. The MHC is a 7-megabase (Mb) region with dense clustering of more than 300 genes that have coordinated roles in immune function.5 With an average gene density of 1/16 kilobases (kb), it is no surprise that more than 420 associations to more than 35 common complex autoimmune, inflammatory, infectious diseases and cancer have been described.6 The classic HLA loci are the best-characterized MHC genes. HLA matching lowers the risks of nonengraftment, acute and chronic graft-versus-host disease (GVHD), and mortality; however, GVHD remains a significant complication in 20% to 80% of patients and points to non-HLA variation as a potential factor.7-11
A comprehensive survey of MHC polymorphisms for potential GVHD determinants requires a complete and precise map of sequence variation at the level of the HLA haplotype, a resource that was made available in 1999 by The MHC Sequencing Consortium12 and was extended through the efforts of The MHC Haplotype Project, The International HapMap Consortium, and The 1000 Genomes Project Consortium.13-18 These resources have provided an unprecedented view of the MHC as a rich source of novel variation that has implications in disease and transplantation. Although the exploration of the MHC in the transplant model is still in the early stages, new information points to the contribution of MHC haplotype–linked variation from both the patient and the donor and enlightens the possible mechanistic pathways through which GVHD arises. In this review, the fundamentals of mapping the MHC for functional variants in unrelated donor HCT are described. Terminology commonly used in HLA immunogenetics and disease mapping are provided in Table 1.
Terms commonly used in HLA immunogenetics and disease mapping
| Term . | Definition . |
|---|---|
| Association mapping | A method for linking clinical characteristics to genetic traits. |
| Block | The physical region of DNA encoding 2 or more SNPs or genes in strong positive LD with one another, such that any one polymorphism is a marker for all of the others in that region. |
| Candidate gene | A gene that has been identified through discovery methods, but not yet validated. |
| Deep sequencing | Sequencing approaches that incorporate multifold coverage of nucleotide positions. |
| Direct sequencing | PCR-based sequencing approaches that use the target DNA strand for amplification and sequencing. |
| Discovery-validation study design | The use of 2 independent cohorts, where one cohort is used to “discover” new trait-associated markers (“candidate” SNPs or genes), and the second cohort is used to “validate” the candidates. |
| Expression quantitative locus | A genetic locus that regulates the level of expression of mRNAs and proteins. |
| Fine mapping | Approaches that use dense SNPs or sequencing to narrow down the region(s) that most likely harbor the causative gene(s). |
| GWAS | The examination of variants across the human genome for association to specific traits or diseases. |
| Genotype | An individual’s DNA sequence (gene, allele, SNP) to be distinguished from phenotype, the trait of the expressed protein, or a clinical characteristic. |
| Haplotype | Markers that travel together on the same chromosome 6 strand (example: HLA-A1-B8-DR3). |
| Heterozygous | Two different HLA alleles/antigens at the HLA locus (example: HLA-A1,2). |
| HLA | HU-1 (human-1 antigen) and LA (leukocyte antigen), antigens that govern tissue type and histocompatibility. |
| HLA class I | The telomeric (further from the centromere) region of the MHC encoding HLA genes. The class I region contains HLA-A, HLA-B, and HLA-C (“classical class I genes”) and HLA-E, F, G; MICA, MICB (“non-classical class I”). |
| HLA class II | The centromeric (closer to the centromere) region of the MHC encoding HLA genes. The class II region contains HLA-DR, HLA-DQ, and HLA-DP. |
| HLA class III | The region of the MHC mapping between class I HLA-B and class II HLA-DR. The class III region contains TNF and the complement pathway genes. |
| Homozygous | Two copies of the same polymorphism or allele (example: SNP genotype AA, TT, CC, or GG; HLA genotype HLA-A*02:01,02:01) |
| Imputation (of SNPs) | Methods that use LD to infer SNPs that have not been directly genotyped. |
| LD | The nonrandom association of alleles or antigens (example: HLA-B8 with HLA-DR3). |
| MHC | A 7.6-Mb region of chromosome 6p21.3 that encodes the histocompatibility genes important in transplantation. |
| Phenotype | A trait. Phenotype can refer to the expressed protein (HLA-A2) or to a clinical characteristic. |
| Proxy | A genetic marker that can be used in lieu of another marker, usually in high LD |
| SNP | The simplest form of human genetic variation. An SNP is a biallelic nucleotide change (example: A vs C). |
| TaqMan | A real-time 5′ nuclease method for genotyping biallelic SNPs. |
| TagSNP | A SNP that is in 100% LD with another SNP(s) so that they serve as proxies for one another. |
| Term . | Definition . |
|---|---|
| Association mapping | A method for linking clinical characteristics to genetic traits. |
| Block | The physical region of DNA encoding 2 or more SNPs or genes in strong positive LD with one another, such that any one polymorphism is a marker for all of the others in that region. |
| Candidate gene | A gene that has been identified through discovery methods, but not yet validated. |
| Deep sequencing | Sequencing approaches that incorporate multifold coverage of nucleotide positions. |
| Direct sequencing | PCR-based sequencing approaches that use the target DNA strand for amplification and sequencing. |
| Discovery-validation study design | The use of 2 independent cohorts, where one cohort is used to “discover” new trait-associated markers (“candidate” SNPs or genes), and the second cohort is used to “validate” the candidates. |
| Expression quantitative locus | A genetic locus that regulates the level of expression of mRNAs and proteins. |
| Fine mapping | Approaches that use dense SNPs or sequencing to narrow down the region(s) that most likely harbor the causative gene(s). |
| GWAS | The examination of variants across the human genome for association to specific traits or diseases. |
| Genotype | An individual’s DNA sequence (gene, allele, SNP) to be distinguished from phenotype, the trait of the expressed protein, or a clinical characteristic. |
| Haplotype | Markers that travel together on the same chromosome 6 strand (example: HLA-A1-B8-DR3). |
| Heterozygous | Two different HLA alleles/antigens at the HLA locus (example: HLA-A1,2). |
| HLA | HU-1 (human-1 antigen) and LA (leukocyte antigen), antigens that govern tissue type and histocompatibility. |
| HLA class I | The telomeric (further from the centromere) region of the MHC encoding HLA genes. The class I region contains HLA-A, HLA-B, and HLA-C (“classical class I genes”) and HLA-E, F, G; MICA, MICB (“non-classical class I”). |
| HLA class II | The centromeric (closer to the centromere) region of the MHC encoding HLA genes. The class II region contains HLA-DR, HLA-DQ, and HLA-DP. |
| HLA class III | The region of the MHC mapping between class I HLA-B and class II HLA-DR. The class III region contains TNF and the complement pathway genes. |
| Homozygous | Two copies of the same polymorphism or allele (example: SNP genotype AA, TT, CC, or GG; HLA genotype HLA-A*02:01,02:01) |
| Imputation (of SNPs) | Methods that use LD to infer SNPs that have not been directly genotyped. |
| LD | The nonrandom association of alleles or antigens (example: HLA-B8 with HLA-DR3). |
| MHC | A 7.6-Mb region of chromosome 6p21.3 that encodes the histocompatibility genes important in transplantation. |
| Phenotype | A trait. Phenotype can refer to the expressed protein (HLA-A2) or to a clinical characteristic. |
| Proxy | A genetic marker that can be used in lieu of another marker, usually in high LD |
| SNP | The simplest form of human genetic variation. An SNP is a biallelic nucleotide change (example: A vs C). |
| TaqMan | A real-time 5′ nuclease method for genotyping biallelic SNPs. |
| TagSNP | A SNP that is in 100% LD with another SNP(s) so that they serve as proxies for one another. |
The genetic landscape of the MHC: coordinated immune function, diversity, and linkage disequilibrium
Three hallmarks of the MHC are the very dense clustering of genes with related immunologic function, extreme sequence diversity of expressed proteins, and strong long-range positive linkage disequilibrium (LD). MHC region gene families include coreceptors (HLA-F, HLA-G), molecules for antigen presentation (26 class I and 24 class II genes), the innate immune response (MICA, MICB, HCP5), inflammation (NKBBiL, LTA, TNF, LTB, LST1, NCR3, AIF1), immune receptors (LY6), heat shock proteins (HSPA1L, HSPA1A, HSPA1B), complement cascade, regulatory receptors (NOTCH4), antigen processing (TAP, HLA-DM, HLA-DO), and peptide transport (RING1). In addition to these gene clusters, the MHC is home to several large families of “housekeeping” genes, including more than 66 histone, 36 zinc finger, and 157 tRNA genes. The coexistence of genes having coordinated immunologic function is thought to reflect some survival advantage to keeping these genes together on the same haplotype.19
The extreme polymorphism of classic HLA genes is part of their role in peptide presentation for host defense against pathogens, and as ligands for natural killer receptors involved in immune surveillance of altered or loss of self-MHC.20 LD describes the nonrandom association of 2 or more markers at frequencies that are higher than would be predicted by chance alone,21 and is the driving force behind the maintenance of alleles on HLA haplotypes. The physical distance over which LD prevails can create exceptionally long MHC haplotypes that measure up to 9 Mb in some populations.22,23 New haplotypes are generated by the shuffling of blocks of conserved sequences at points of recombination.19
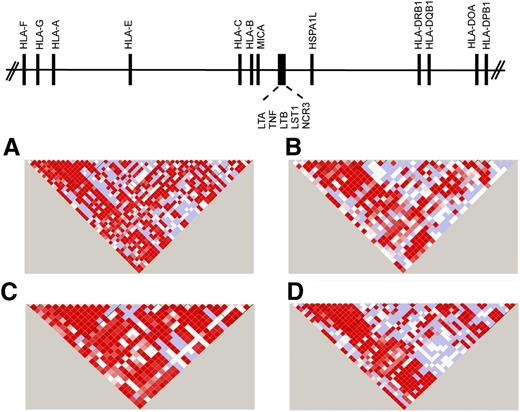
The first complete sequence of the MHC was derived from multiple different HLA haplotypes.12 Subsequently, sequencing of cell lines homozygous at HLA loci provided haplotype-specific information on common and rare variants, information that has been instrumental for fine mapping.13-16,24-28 The complete sequencing of HLA haplotypes shows that the MHC is a microcosm of the human genome, encoding single nucleotide polymorphisms (SNPs), substitutions and deletion-insertion polymorphisms, short tandem repeats (microsatellites), and copy number variations. The simplest form is the SNP, which is a powerful tool for surveying regions containing disease-associated genes.27,29-34 Strong positive LD among SNPs gives rise to “blocks” of short SNP haplotypes that can be readily identified by genotyping 1 representative “tag SNP” for the block. TagSNPs that have portability across ethnically diverse populations are particularly powerful tools for mapping disease-susceptibility genes35-37 (Figure 138,39 ).
SNP haplotypes in white, African, Asian, and admixed populations from the 1000 Genomes. Continental populations may differ for the minor allele frequencies of SNPs, the strength of LD, and recombination, factors that define haplotype block structure within the MHC. Illustrated is the 20.8-kb TNF block consisting of the LTA-TNF-LST-NCR loci for samples from the 1000 Genomes Project Consortium18,38 for individuals of diverse backgrounds. (A) White European, n = 379; (B) African, n = 246; (C) Asian, n = 286; and (D) admixed American (includes self-described Hispanic), n = 181. The intensity of the red color in each pairwise SNP comparison is a measure of the strength of LD as measured by D′.39 Bright red boxes indicate a D′ of 1 (100% linkage), and the gradation of color to white indicates progressively smaller D′, values indicative of weaker LD. The different block structure and SNP haplotypes among these ethnically diverse populations may facilitate the fine mapping of SNP association signals within this region of the MHC.
SNP haplotypes in white, African, Asian, and admixed populations from the 1000 Genomes. Continental populations may differ for the minor allele frequencies of SNPs, the strength of LD, and recombination, factors that define haplotype block structure within the MHC. Illustrated is the 20.8-kb TNF block consisting of the LTA-TNF-LST-NCR loci for samples from the 1000 Genomes Project Consortium18,38 for individuals of diverse backgrounds. (A) White European, n = 379; (B) African, n = 246; (C) Asian, n = 286; and (D) admixed American (includes self-described Hispanic), n = 181. The intensity of the red color in each pairwise SNP comparison is a measure of the strength of LD as measured by D′.39 Bright red boxes indicate a D′ of 1 (100% linkage), and the gradation of color to white indicates progressively smaller D′, values indicative of weaker LD. The different block structure and SNP haplotypes among these ethnically diverse populations may facilitate the fine mapping of SNP association signals within this region of the MHC.
The MHC and GVHD
The MHC is a prime region of interest for the exploration of novel GVHD determinants because of the density of genes with immune function that currently are not tested in routine clinical practice.11,40,41 Untyped patient and donor variation responsible for GVHD can now be surveyed with great precision with the availability of SNP arrays. The identification of GVHD determinants is performed using association mapping techniques. Candidate gene studies focus on specific genes that have a plausible basis to the disease. Genome-wide association studies (GWAS) test associations of SNPs to clinical endpoints without a priori assumptions about the nature of the gene(s) involved in disease pathogenesis. Both approaches provide important information on the genes and allele variants that may be clinically relevant in transplantation. Candidate gene approaches may permit genes with known putative function to be analyzed individually or together with other candidate genes to assess the effects of gene-gene interactions that may affect the same biological process or clinical end point. GWAS approaches, on the other hand, permit novel, heretofore unknown, genes and pathways to be discovered, which can then be further studied in directed analyses of specific genes or pathways. The choice of method is largely driven by whether there are specific mechanistic hypotheses that can best be addressed through the study of specific candidate genes, or whether the research question is to broadly explore and identify genome-wide factors, variants that have previously been characterized in candidate gene studies, as well as novel polymorphisms. Both approaches require in vitro or in vivo functional studies to ultimately define how the sequence variation affects gene function and how the polymorphism may lead to disease.
The number of candidate gene studies in HCT far exceeds that for GWAS. Among candidate gene studies, the heterogeneity of results has prompted recent analyses of independent cohorts.42 Many factors could influence the associations of a given variant to a clinical end point, including patient and donor characteristics such as age, sex, parity of female individuals, race and ethnicity, and so on. In addition, studies may differ with respect to the intensity of the conditioning regimen, the use of T-cell depletion for GVHD prevention, and posttransplant cellular therapy. Hence, interpretation of the results of retrospective genetic association studies ideally includes careful consideration of the characteristics of the study population. Equally important for candidate gene studies is the oftentimes lack of uniformity of the specific polymorphisms within the candidate gene that are genotyped. For example, a regulatory region of interest may be polymorphic, with several different sites of variation; when different sites are typed and analyzed, the conclusions may or may not pertain to the untyped sites. These methodologic differences add to the complexity of interpreting results across studies.
Candidate MHC gene studies in HCT
The most extensively studied non-HLA MHC genes to date are involved in inflammation, the stress response, innate immunity, and natural killer (NK)–T-cell allorecognition (Table 2): tumor necrosis factor (TNF)-α,43-49 MICA,50-52 HLA-E,48,53-58 HLA-G,59-61 and HSPA1L.62
Summary of MHC region polymorphisms associated with clinical outcome after allogeneic HCT
| A. Candidate gene/polymorphism . | Model . | Study population . | Effect . | Reference . | |
|---|---|---|---|---|---|
| HLA-G | 14-bp insertion/deletion in 3′ UTR | Homozygosity for del/del | 53 HLA-matched URD | Increased acute GVHD | 59 |
| Homozygosity for ins/ins | 47 HLA-matched URD, siblings, cord blood | Lower survival and DFS | 61 | ||
| 157 HLA-identical siblings | Increased severe acute GVHD | 60 | |||
| HLA-E | E*01:01 and E*01:03 alleles | Patient and donor homozygosity for E*01:03 | 187 HLA-identical siblings (ref 54); 33 HLA-identical siblings, 12 HLA-matched URD; 11 haploidentical-mismatched related donors (ref 57) | Lower grades 3-4 acute GVHD; lower TRM; improved survival | 54,57 |
| Patient E*01:03 homozygosity | 67 HLA-matched siblings; 16 HLA matched URD | Higher survival; higher DFS; lower TRM | 55 | ||
| Donor E*01:03 homozygosity | 124 HLA-matched URD | Lower grades 2-4 acute GVHD; lower chronic GVHD; higher relapse | 56 | ||
| 116 HLA-matched URD | No correlation with clinical outcome | 58 | |||
| E*01:01/01:03 | Mismatch | 116 HLA-matched URD | No correlation with clinical outcome | 58 | |
| Rs1264457 residue 128 arg (R) vs gly (G) | Mismatch | 922 HLA-matched and mismatched URD | Lower survival | 48 | |
| MICA | Residue 129 valine (V) versus methionine (M) | Patients homozygous for 129V | 211 HLA-matched siblings | Higher chronic GVHD | 50 |
| Mismatch | 236 HLA-matched and mismatched URD | Higher grades 2-4 acute GVHD, notably gastrointestinal GVHD | 51 | ||
| 38 HLA-matched URD | No association | 52 | |||
| Binding affinity for M129V | 38 HLA-matched URD | No association | 52 | ||
| MICA*008 | Patient genotype | 38 HLA-matched URD | Lower acute GVHD | 52 | |
| MICA*004, 006, 009, 044, 049. | Patient genotype | 38 HLA-matched URD | Higher gastrointestinal acute GVHD | 52 | |
| TNF | Rs1799964 (-1031) | Mismatch | 922 HLA-matched and mismatched URD | Higher grade 4 acute GVHD | 48 |
| Patient C genotype | 180 HLA-matched URD | Higher grades 3-4 acute GVHD | 47 | ||
| C genotype | 182 HLA-matched and mismatched URD | Higher TRM | 45 | ||
| Patient genotype, donor genotype | 612 HLA-matched related donors; 686 HLA-matched URD | No correlation | 42 | ||
| −1031/-863/-857 (rs1799724) U02/U03 alleles | Patients and/or donors with U02 and/or U03 alleles | 462 HLA-matched and mismatched URD | Higher grades 3-4 acute GVHD and lower relapse | 43 | |
| −308 | Genotype | 111 HLA-matched URD; 85 HLA matched siblings | No correlation | 44 | |
| Rs1800630 (-863) d4/d4 allele | Patient genotype | 180 HLA-matched URD | Higher grades 3-4 acute GVHD | 47 | |
| 111 HLA-matched URD; 85 HLA matched siblings | Higher grades 2-4 acute GVHD | 44 | |||
| D4/d5 microsatellite alleles | Donor alleles | 246 HLA-matched URD | Higher mortality | 49 | |
| Rs1800630* | Patient genotype, donor genotype | 612 HLA-matched related donors; 686 HLA-matched URD | No correlation | 42 | |
| RS1800629* | Patient genotype, donor genotype | 612 HLA-matched relateds; 686 HLA-matched URD | No correlation | 42 | |
| HSPA1L | +2763G/A | Patients homozygous for AA genotype | 63 HLA-matched siblings; 70 haploidentical related and HLA-matched URD | Higher grades 2-4 acute GVHD; no correlation with donor HSPA1L or patient-donor match/mismatch | 62 |
| Rs2075800 (see section C below) | Genotype; mismatch | 922 HLA-matched and mismatched URD | No correlation | 48 | |
| B. GWAS chromosome 6 signals/closest locus | Model | Study population | Effect | Reference | |
| Rs6937034 | HLA-DP | Mismatch | 1598 HLA-matched URD | Higher grades 2-4 acute GVHD | 63 |
| C. “MHC-was”/Closest Locus | Model | Study Population | Effect | Reference | |
| Rs209130‡ | TRIM27 | Mismatch | 2628 HLA-mismatched URD | Higher grades 3-4 acute GVHD | 41 |
| rs2523957‡ | HLA-A | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Higher chronic GVHD | 41 |
| rs887464† | PSOR1C3/POU5F1 | Mismatch (patient-versus-donor) | 4205 HLA-matched URD | Lower survival | 40 |
| rs2244546‡ | HCP5 | Donor genotype GG | 2628 HLA-mismatched URD | Higher relapse | 41 |
| rs915654‡ | LTA | Patient genotype AT | 2628 HLA-mismatched URD | Lower TRM | 41 |
| Rs2242656‡ | BAG6 | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Higher grades 2-4 acute GVHD | 41 |
| rs2075800‡ | HSPA1L | Patient genotype GG | 2628 HLA-mismatched URD | Higher grades 3-4 acute GVHD | 41 |
| rs3830076‡ | FKBPL | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Lower chronic GVHD | 41 |
| rs394657‡ | NOTCH4 | Donor genotype AA | 2628 HLA-mismatched URD | Lower grades 3-4 acute GVHD | 41 |
| rs2071479‡ | HLA-DOB | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Higher chronic GVHD | 41 |
| rs429916‡ | HLA-DOA | Patient genotype AA | 2628 HLA-mismatched URD | Lower survival | 41 |
| Rs2281389† | HLA-DPB1 3′UTR | Mismatch | 4205 HLA-matched URD | Higher grades 2-4 acute GVHD | 40 |
| rs986522‡ | COL11A2 | Donor genotype CG | 2628 HLA-mismatched URD | Lower relapse | 41 |
| rs107822‡ | RING1A | Mismatch | 2628 HLA-mismatched URD | Higher chronic GVHD | 41 |
| A. Candidate gene/polymorphism . | Model . | Study population . | Effect . | Reference . | |
|---|---|---|---|---|---|
| HLA-G | 14-bp insertion/deletion in 3′ UTR | Homozygosity for del/del | 53 HLA-matched URD | Increased acute GVHD | 59 |
| Homozygosity for ins/ins | 47 HLA-matched URD, siblings, cord blood | Lower survival and DFS | 61 | ||
| 157 HLA-identical siblings | Increased severe acute GVHD | 60 | |||
| HLA-E | E*01:01 and E*01:03 alleles | Patient and donor homozygosity for E*01:03 | 187 HLA-identical siblings (ref 54); 33 HLA-identical siblings, 12 HLA-matched URD; 11 haploidentical-mismatched related donors (ref 57) | Lower grades 3-4 acute GVHD; lower TRM; improved survival | 54,57 |
| Patient E*01:03 homozygosity | 67 HLA-matched siblings; 16 HLA matched URD | Higher survival; higher DFS; lower TRM | 55 | ||
| Donor E*01:03 homozygosity | 124 HLA-matched URD | Lower grades 2-4 acute GVHD; lower chronic GVHD; higher relapse | 56 | ||
| 116 HLA-matched URD | No correlation with clinical outcome | 58 | |||
| E*01:01/01:03 | Mismatch | 116 HLA-matched URD | No correlation with clinical outcome | 58 | |
| Rs1264457 residue 128 arg (R) vs gly (G) | Mismatch | 922 HLA-matched and mismatched URD | Lower survival | 48 | |
| MICA | Residue 129 valine (V) versus methionine (M) | Patients homozygous for 129V | 211 HLA-matched siblings | Higher chronic GVHD | 50 |
| Mismatch | 236 HLA-matched and mismatched URD | Higher grades 2-4 acute GVHD, notably gastrointestinal GVHD | 51 | ||
| 38 HLA-matched URD | No association | 52 | |||
| Binding affinity for M129V | 38 HLA-matched URD | No association | 52 | ||
| MICA*008 | Patient genotype | 38 HLA-matched URD | Lower acute GVHD | 52 | |
| MICA*004, 006, 009, 044, 049. | Patient genotype | 38 HLA-matched URD | Higher gastrointestinal acute GVHD | 52 | |
| TNF | Rs1799964 (-1031) | Mismatch | 922 HLA-matched and mismatched URD | Higher grade 4 acute GVHD | 48 |
| Patient C genotype | 180 HLA-matched URD | Higher grades 3-4 acute GVHD | 47 | ||
| C genotype | 182 HLA-matched and mismatched URD | Higher TRM | 45 | ||
| Patient genotype, donor genotype | 612 HLA-matched related donors; 686 HLA-matched URD | No correlation | 42 | ||
| −1031/-863/-857 (rs1799724) U02/U03 alleles | Patients and/or donors with U02 and/or U03 alleles | 462 HLA-matched and mismatched URD | Higher grades 3-4 acute GVHD and lower relapse | 43 | |
| −308 | Genotype | 111 HLA-matched URD; 85 HLA matched siblings | No correlation | 44 | |
| Rs1800630 (-863) d4/d4 allele | Patient genotype | 180 HLA-matched URD | Higher grades 3-4 acute GVHD | 47 | |
| 111 HLA-matched URD; 85 HLA matched siblings | Higher grades 2-4 acute GVHD | 44 | |||
| D4/d5 microsatellite alleles | Donor alleles | 246 HLA-matched URD | Higher mortality | 49 | |
| Rs1800630* | Patient genotype, donor genotype | 612 HLA-matched related donors; 686 HLA-matched URD | No correlation | 42 | |
| RS1800629* | Patient genotype, donor genotype | 612 HLA-matched relateds; 686 HLA-matched URD | No correlation | 42 | |
| HSPA1L | +2763G/A | Patients homozygous for AA genotype | 63 HLA-matched siblings; 70 haploidentical related and HLA-matched URD | Higher grades 2-4 acute GVHD; no correlation with donor HSPA1L or patient-donor match/mismatch | 62 |
| Rs2075800 (see section C below) | Genotype; mismatch | 922 HLA-matched and mismatched URD | No correlation | 48 | |
| B. GWAS chromosome 6 signals/closest locus | Model | Study population | Effect | Reference | |
| Rs6937034 | HLA-DP | Mismatch | 1598 HLA-matched URD | Higher grades 2-4 acute GVHD | 63 |
| C. “MHC-was”/Closest Locus | Model | Study Population | Effect | Reference | |
| Rs209130‡ | TRIM27 | Mismatch | 2628 HLA-mismatched URD | Higher grades 3-4 acute GVHD | 41 |
| rs2523957‡ | HLA-A | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Higher chronic GVHD | 41 |
| rs887464† | PSOR1C3/POU5F1 | Mismatch (patient-versus-donor) | 4205 HLA-matched URD | Lower survival | 40 |
| rs2244546‡ | HCP5 | Donor genotype GG | 2628 HLA-mismatched URD | Higher relapse | 41 |
| rs915654‡ | LTA | Patient genotype AT | 2628 HLA-mismatched URD | Lower TRM | 41 |
| Rs2242656‡ | BAG6 | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Higher grades 2-4 acute GVHD | 41 |
| rs2075800‡ | HSPA1L | Patient genotype GG | 2628 HLA-mismatched URD | Higher grades 3-4 acute GVHD | 41 |
| rs3830076‡ | FKBPL | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Lower chronic GVHD | 41 |
| rs394657‡ | NOTCH4 | Donor genotype AA | 2628 HLA-mismatched URD | Lower grades 3-4 acute GVHD | 41 |
| rs2071479‡ | HLA-DOB | Mismatch (donor-versus-patient) | 2628 HLA-mismatched URD | Higher chronic GVHD | 41 |
| rs429916‡ | HLA-DOA | Patient genotype AA | 2628 HLA-mismatched URD | Lower survival | 41 |
| Rs2281389† | HLA-DPB1 3′UTR | Mismatch | 4205 HLA-matched URD | Higher grades 2-4 acute GVHD | 40 |
| rs986522‡ | COL11A2 | Donor genotype CG | 2628 HLA-mismatched URD | Lower relapse | 41 |
| rs107822‡ | RING1A | Mismatch | 2628 HLA-mismatched URD | Higher chronic GVHD | 41 |
Part A: Candidate gene studies. In these studies, the specific MHC region gene or allele or SNP was tested individually. Genes are presented in chromosomal order. Part B: MHC region SNPs identified by GWAS of the entire genome. In this study, GWAS was performed using a commercial platform. Part C: SNPs identified by “MHC-was” SNPs survey. In these studies, SNPs genotyping was performed using commercial platforms. SNPs are presented in chromosomal order.
del, deletion; DFS, disease-free survival; ins, insertion.
SNP was imputed.42
SNP was validated in a discovery-validation study design.40
SNP was identified in a discovery set; validation is pending.41
TNF-α
The TNF block is a 7-kb region within class III that encodes TNF-α, TNF-β, lymphocytoxin (LTA)-α, leukocyte-specific transcript (LST)-1, and allograft inflammatory factor (AIF).28,64,65 These genes share coordinated function as inflammatory mediators.66 TNF-α has been the most comprehensively studied candidate gene,43-49 and its sequence diversity and haplotype organization have been defined.48,49,67,68
TNF-α is a proinflammatory Th1 cytokine involved in different phases of GVHD.69 In phase 1, TNF-α is associated with tissue injury and the initiation of the cytokine storm; the activation of antigen-presenting cells enhances alloantigen presentation and induces inflammatory cytokines, leading to the recruitment of the effector cells that ultimately mediate damage of the target organs. In phase 3, TNF-α and IL1 participate in the cytokine storm.69 The TNF-α sequence variation is associated with GVHD, transplant-related mortality (TRM), and survival (Table 2).43,45-49,70 Sequence variation at the −308 (A/G) and the −863 (A/C) positions in the 5′ promoter account for differential expression of TNF-α, which correlate with the risk of acute GVHD.71
MICA
MICA is polymorphic,72 has strong haplotypic associations to HLA-B,73,74 is inducible by stress,75 and is a ligand for activating NKG2D receptors.76 A potential role for MICA in GVHD might involve T cell- or NK-mediated pathways and/or inflammatory and stress mediators. One of the earliest investigations of the MICA region demonstrated the association of block-matching with improved survival after HCT,77 paving the way for additional studies of this locus.50-52 Methionine (M) or valine (V) at residue 129 correlates with the strength of binding of MICA to its NKG2D receptor.78 Patients with 129V alleles have an increased risk of chronic GVHD after sibling donor HCT.50 Furthermore, patient-donor MICA mismatching has correlated with an increased risk of II-IV acute GVHD, particularly of the gastrointestinal tract after HLA-matched and HLA-mismatched HCT.51 In HLA-matched unrelated donor HCT, MICA*008-positive patients had less acute GVHD.52 These data collectively suggest that the presence of specific MICA phenotypes may serve as antigenic targets that lead to GVHD or that influence the interaction of MICA to its receptors.
HLA-E
HLA-E plays a dual role in innate and adaptive immunity by functioning as a ligand for inhibitory CD94/NKG2A receptors and by presenting peptides. The 2 major alleles, E*01:01 and 01:03, differ from one another at residue 107 (glycine [G] vs arginine [R]) of the α2 heavy chain.72 HLA-E alleles have different levels of cell surface expression, with HLA-E*01:01 expressed at lower levels than E*01:03.79 Homozygosity for E*01:03 was associated with a lower risk of acute GVHD and TRM,54,57 and lower TRM and improved survival.55 In other studies, associations of HLA-E with acute GVHD have been heterogeneous.56,58 Patient-donor mismatching for the G107R substitution defined by rs126457 is associated with lower survival.48
HLA-G
This nonclassic class I gene has several unique features including a 14-bp insertion or deletion located in its 3′ untranslated region (UTR) and at least 15 isoforms that are generated through alternative splicing.72,80-82 Because of its multiple allotypes, HLA-G has been studied as a potential source of donor-recipient disparity in transplantation. The 14-bp insertion/deletion was associated with acute GVHD in some,59,60 but not other, studies,61 although homozygosity for the 14-bp deletion did correlate with lower survival and disease-free survival.
GWAS
GWAS studies of GVHD in transplantation ideally require upwards of 4000 transplants for sufficient power across a range of odds ratios.83 GWAS of HLA-matched sibling or unrelated donor transplant populations permit the risks associated with nonchromosome 6 genes to be measured without the effects of HLA mismatching. However, HLA-mismatched populations provide unique information on the additive or synergistic effects of HLA mismatching with variation, both within and outside of the MHC. The Japan Marrow Donor Program has conducted the largest GWAS analysis of transplant outcomes to date.63 Designed as a 2-stage analysis, the first stage assumed no HLA restriction of SNPs and identified rs6937034 near HLA-DP as a marker of grades 2-4 acute GVHD. The second phase tested the hypothesis that SNPs function as HLA-restricted minor histocompatibility antigens; rs17473423 (chromosome 12) associated with risk of grades 3-4 acute GVHD in HLA-A*24-B*52-C*12-DRB1*15-DQB1*06–positive transplants. Furthermore, rs9657655 (chromosome 9) was a marker for HLA-A*33:03-B*44:03-C*14:03–positive transplants, and 4 additional loci were identified in association with HLA-DQB1*05:01, C*01:02, B*52:01, and C*12:02. These data suggest a role for non-HLA polymorphisms as minor histocompatibility antigens restricted by HLA on extended Japanese HLA haplotypes. The investigators further identified 2 SNPs associated with patient genotype (chromosome 12 rs5998746 and chromosome 18 rs11873016); no GVHD-associated SNPs were identified in the donor genotype model.
MHC region SNP discovery
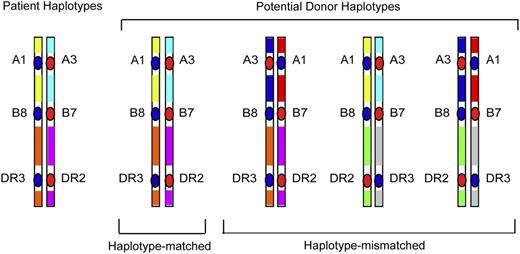
Whereas GWAS studies survey polymorphisms across the entire human genome, a focused examination of dense SNPs within the MHC provides a firsthand look at untyped variation carried on HLA haplotypes of the patient and donor. As described before, the rationale for such “MHC-was” studies is based on the concept that unrelated individuals with the same tissue type may have different HLA haplotypes that give rise to intergenic sequence variation (Figure 2), including very common HLA haplotypes (Figure 3). Focused SNP mapping of the MHC is applicable not only to HLA-mismatched but also to HLA-matched transplant pairs.11,40,41 As with GWAS, there are no a priori regions or genes that are more or less likely to cause GVHD.
HLA-mismatched patient-unrelated donors may encode variation in regions between classical HLA loci. HLA-matched patients and donors may encode different sequences in between the HLA-A, C, B, DRB1, and DQB1 loci, and these variants may be a source of disparity that gives rise to GVHD after HCT.40 In HLA-mismatched HCT, the frequency of patient-donor sequence differences and the extent of the MHC that is different may be pronounced, and the effects of such untyped variation may be additive to that from the patient-donor HLA mismatch.41
HLA-mismatched patient-unrelated donors may encode variation in regions between classical HLA loci. HLA-matched patients and donors may encode different sequences in between the HLA-A, C, B, DRB1, and DQB1 loci, and these variants may be a source of disparity that gives rise to GVHD after HCT.40 In HLA-mismatched HCT, the frequency of patient-donor sequence differences and the extent of the MHC that is different may be pronounced, and the effects of such untyped variation may be additive to that from the patient-donor HLA mismatch.41
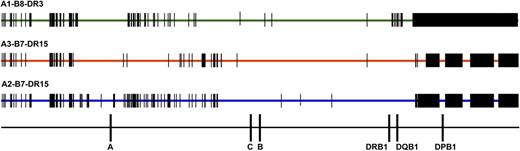
Comparison of SNPs encoded on HLA-A1-B8-DR3, HLA-A3-B7-DR15, and HLA-A2-B7-DR15 haplotypes. SNP variation encoded on the 3 most common white haplotypes—HLA-A1-B8-DR3, A3-B7-DR15, and A2-B7-DR15—are a source of patient-donor variation. Data are shown for 28 HLA-A1-B8-DR3 chromosomes, 4 HLA-A3-B7-DR15 chromosomes, and 4 HLA-A2-B7-DR15 chromosomes. HLA-A1-B8-DR3 chromosomes share conserved sequences, indicated by the green bar; SNP substitutions are indicated by vertical black bars and black rectangles. HLA-matched and HLA-mismatched patients and donors may differ at SNP positions across the 7 Mb of the MHC. For example, a patient with the HLA-A1,3-B7,8-DR15,3 tissue type and donor with the HLA-A1,2-B7,8-DR15,3 tissue type (HLA-A mismatched) will have disparities at multiple nucleotide positions. These untyped SNPs may be a source of variation associated with increased risk of GVHD and TRM.41
Comparison of SNPs encoded on HLA-A1-B8-DR3, HLA-A3-B7-DR15, and HLA-A2-B7-DR15 haplotypes. SNP variation encoded on the 3 most common white haplotypes—HLA-A1-B8-DR3, A3-B7-DR15, and A2-B7-DR15—are a source of patient-donor variation. Data are shown for 28 HLA-A1-B8-DR3 chromosomes, 4 HLA-A3-B7-DR15 chromosomes, and 4 HLA-A2-B7-DR15 chromosomes. HLA-A1-B8-DR3 chromosomes share conserved sequences, indicated by the green bar; SNP substitutions are indicated by vertical black bars and black rectangles. HLA-matched and HLA-mismatched patients and donors may differ at SNP positions across the 7 Mb of the MHC. For example, a patient with the HLA-A1,3-B7,8-DR15,3 tissue type and donor with the HLA-A1,2-B7,8-DR15,3 tissue type (HLA-A mismatched) will have disparities at multiple nucleotide positions. These untyped SNPs may be a source of variation associated with increased risk of GVHD and TRM.41
SNPs are proxies for genes that cause disease. In transplantation, risks might be conferred by patient genotypes, donor genotypes, and/or patient-donor mismatching. The patient and the donor genotype models test the hypothesis that the presence of specific variants affects GVHD, relapse, or survival. This concept is akin to classical disease-association mapping.84 SNPs may be proxies for regulatory variants that affect the transcription and translation of the gene or reside a distance away from the target gene(s), including genes that reside on other chromosomes. Alternatively, SNPs may result in nonsynonymous coding substitutions that affect the structure and function of proteins, ligand-receptor interactions, or possible interactions with other molecules in shared pathways. The most conceptually straightforward model is that of patient-donor mismatching, in which the patient’s SNP allele differs from the donor’s. This model indirectly tests whether there are donor polymorphisms recognized by the patient (donor AG vs patient AA or GG), patient variation recognized by the donor (patient AG vs donor AA or GG), or both (patient AA vs donor GG). The identification of any SNP in any model sheds light on what candidate gene(s) are involved in GVHD, and the potential functional consequences of DNA variation.
Two recent studies demonstrate that the MHC harbors clinically relevant variation responsible for GVHD and support the concept that GVHD is a polygenic disease.40,41 A discovery-validation study of more than 4200 HLA-A,C,B,DRB1,DQB1–matched (“10/10”) unrelated donor transplants identified 2 SNPs associated with clinical outcome.40 Donor mismatching at rs887464 (donor AG vs patient AA) was associated with lower survival and disease-free survival. Interestingly, rs887464 is a risk marker for type 1 diabetes and a putative expression quantitative locus.84-86 These data suggest that GVHD may share common pathways with other autoimmune diseases and offer new approaches for fine mapping the gene(s) that are involved in GVHD.87
In HLA-matched unrelated donor HCT, patient-donor mismatching at rs2281389 (patient AG vs donor GG) increased the risks of grades 2-4 acute GVHD.40 SNP rs2281389 resides centromeric to the HLA-DPB1 3′ UTR and shows complex haplotype linkage to HLA-DP; its biological implications have yet to be elucidated. Both rs887464 and rs2281389 were identified in the mismatch model, suggesting that HLA-matched patients and donors have untyped disparity outside of the classical HLA loci that can increase the risks of GVHD and mortality.
The evaluation of HLA-mismatched unrelated donor HCT introduces different challenges than the HLA-matched setting. The hypothesis that deleterious HLA haplotype–linked polymorphisms can synergize with HLA mismatches, per se, has recently been tested in a retrospective study of 2628 transplants with a single HLA-A,C,B,DRB1, or DQB1 (“9/10”) mismatch.41 Untyped SNPs “hitchhike” with the HLA alleles and the rate of SNP mismatching may be particularly high near the HLA-mismatched locus. A survey of more than 1100 SNPs identified 12 candidate markers for future validation. Risks of acute and chronic GVHD, relapse, and mortality increased with increasing numbers of unfavorable SNPs, and parallel observations from GWAS mapping of complex autoimmune diseases.87 Finally, SNPs define HLA-SNP haplotypes. The most common white haplotype worldwide, HLA- A*01:01-B*08:01-DRB1*03:01 (7.4% frequency88 ), displays variation at rs429916, a marker for survival.41 These data suggest that knowledge of the patient’s and donor’s haplotypes not only for the specific HLA alleles, but also haplotype-linked SNPs, are needed to fully understand the risks to individual patients after HCT. Future studies of SNPs and HLA haplotypes will be needed to define the extent to which the SNP-HLA haplotypes are shared among ethnically diverse populations and those that are unique.
Impact of HLA mismatching
If risks can stem from non-HLA variants that are carried on HLA haplotypes, then cumulative effects of both HLA and non-HLA variation may be significant. To test this hypothesis, the risks associated with a single HLA-A, B, DRB1, DQB1 allele or antigen, or single HLA-C allele mismatch relative to a single HLA-C antigen mismatch were defined in models that adjusted for the contribution of the 12 SNP genotypes or mismatching.41 The risks associated with allele or antigen mismatching at each locus were similar to HLA-C antigen mismatches, with the exception of HLA-DQB1 mismatches, which as a group were less risky. Although these data suggest that, in general, any HLA mismatch outside of HLA-DQ is deleterious, some HLA mismatch combinations are more likely to encode favorable SNPs than others. It is intriguing to speculate that the more favorable outcomes of HLA-C allele mismatches might be contributed partly by the hitchhiking of favorable SNPs on the patient’s and donor’s haplotypes. That the permissiveness of HLA mismatches might be defined by synergistic effects between HLA coding region variation and haplotype-linked variation introduces a new hypothesis that will require large cohorts of SNP-typed transplants for testing in the future.
Comparison of HLA 10/10 with 9/10 unrelated donor HCT
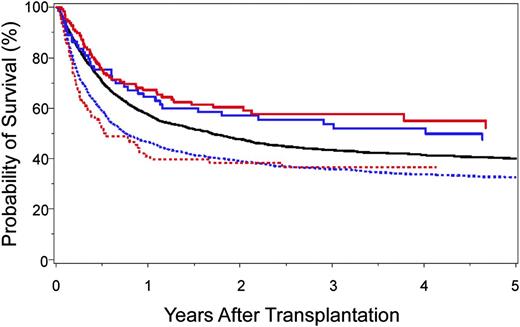
The identification of clinically relevant MHC region SNPs permits a formal comparison of survival after 10/10-matched and 9/10-matched unrelated donor HCT (Figure 4). Survival after 9/10 HCT with 0 to 1 unfavorable SNP is similar to that after 10/10 HCT with favorable SNPs,40,41 and both are superior to transplantation from 9/10 donors with 2 or more unfavorable SNPs or from 10/10 donors with an unfavorable SNP. Intermediate risks are observed after 10/10 transplantation with a “neutral” SNP.40 These data shed new light on clinical settings, where the use of a 9/10 donor may yield comparable outcomes with that of a 10/10 donor and may suggest that prospective patient and donor evaluation for SNPs may aid the selection of donors with optimal MHC SNPs.
Kaplan-Meier probability of survival after HLA 10/10-matched and HLA 9/10-matched unrelated donor HCT according to the number of favorable SNPs. A total of 4205 HLA 10/10 allele-matched40 and 2628 HLA 9/10 single locus–mismatched41 unrelated donor-patient pairs were examined for SNP associations with survival: 10/10-matched and favorable rs887464 (SNP defined in reference 40) (solid red curve; n = 126); 9/10-matched and 0–1 unfavorable SNP (SNPs defined in reference 41) (solid blue curve; n = 73); 10/10-matched and neutral rs887464 (solid black curve; n = 3521); 9/10-matched and more than 1 unfavorable SNP (hatched blue curve; n = 1849); and 10/10-matched and unfavorable rs887464 (hatched red curve; n = 92). Overall survival after 10/10-matched unrelated donor HCT with favorable SNP is similar to that after 9/10-matched unrelated HCT with 0–1 unfavorable SNP. The negative effect of cumulative unfavorable SNPs is additive to HLA mismatch–associated risks.40,41
Kaplan-Meier probability of survival after HLA 10/10-matched and HLA 9/10-matched unrelated donor HCT according to the number of favorable SNPs. A total of 4205 HLA 10/10 allele-matched40 and 2628 HLA 9/10 single locus–mismatched41 unrelated donor-patient pairs were examined for SNP associations with survival: 10/10-matched and favorable rs887464 (SNP defined in reference 40) (solid red curve; n = 126); 9/10-matched and 0–1 unfavorable SNP (SNPs defined in reference 41) (solid blue curve; n = 73); 10/10-matched and neutral rs887464 (solid black curve; n = 3521); 9/10-matched and more than 1 unfavorable SNP (hatched blue curve; n = 1849); and 10/10-matched and unfavorable rs887464 (hatched red curve; n = 92). Overall survival after 10/10-matched unrelated donor HCT with favorable SNP is similar to that after 9/10-matched unrelated HCT with 0–1 unfavorable SNP. The negative effect of cumulative unfavorable SNPs is additive to HLA mismatch–associated risks.40,41
After SNP discovery: replication and fine mapping
Candidate SNPs that are replicated for the same model (genotype or mismatch) and for the same clinical endpoint (GVHD, relapse, survival) represent bone fide tags for the causative genes.40,89,90 Fine mapping involves focused studies to narrow down the regions harboring the causative gene(s) for comprehensive analysis of sequence variation. Often, the physical proximity of the SNP to a gene is used to initiate direct examination of gene polymorphism and function. When the SNP itself defines a nonsynonymous substitution, the protein-coding gene is a prime suspect. In gene-dense regions, sequencing several hundred kb 5′ and 3′ of the target gene may be necessary, depending on local block structure.
Often the SNP associations have biological plausibility because they define known genes with known immunologic function. In some instances, the SNP may be shared among different diseases; for example, Crohn’s colitis, multiple sclerosis, psoriasis, rheumatoid arthritis, systemic lupus erythematosus, and type 1 diabetes mellitus share as much as 44% of SNP associations.91 Shared SNPs provide an approach for fine mapping across different diseases and phenotypes.84,85
Dense SNP panels and “deep sequencing” with next-generation sequencing are two of the most common methods used for fine mapping. New genome-wide approaches that facilitate the mapping of SNPs to structural alleles offer promising methods for analyzing many genomes concurrently.92 When haplotype block structure varies among populations,35-37,87,93 SNPs that are shared among different haplotypes may be helpful for ruling in or ruling out candidate genes (Figure 1). In some instances, it may not be feasible to identify a single causative variant, but rather haplotypes of tightly-linked markers.
Future applications to clinical practice
Mapping GVHD-associated SNPs in a retrospective population of paired patients and donors provides information on the frequency of SNPs among donors who were selected for transplantation using criteria other than SNPs. The likelihood that a given patient has HLA-matched donors who also have favorable SNPs can be estimated by examining SNP frequencies in a pool of individuals who share the same tissue type. In a retrospective study of 230 patients referred to a single center for initiation of an unrelated donor search,40 patients for whom at least 2 HLA-matched donors were identified were each genotyped for the GVHD-associated rs2281389. Among 131 patients with 2 donors each, 82% had at least one rs2281389-matched donor and 48% had more than one SNP-matched donor. Among 17 patients with 6 HLA-matched donors each, 94% had at least 1 rs2281389-matched donor and 88% had more than 1 SNP-matched donor. These data suggest that rs2281389-matched donors can be identified among HLA-matched donors, and that the likelihood of finding such donors increases with more donors tested per patient. Similar frequency data are needed in the future for validated SNPs, because the frequency of donors with favorable SNPs may depend on their haplotypes.41
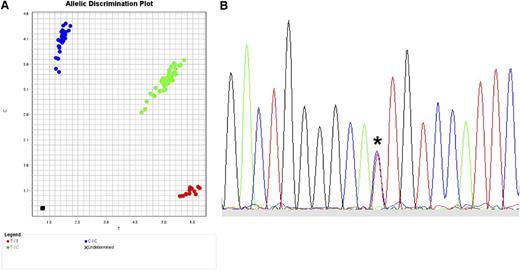
The translation of SNP data to clinical practice in the future will depend on what kind of polymorphism should be tested, whether quantitative protein expression is a robust indicator of GVHD risk, and whether tissue-specific patterns of expression are clinically relevant. The most straightforward translation of validated SNPs to the clinical setting requires efficient and highly robust read-out assays. One available method for SNP genotyping is the TaqMan 5′ nuclease allelic discrimination assay, an efficient, low-cost method in which data interpretation is unambiguous because only 2 alleles are possible (Figure 5A).29,94,95 Direct sequencing is also feasible and useful for selected applications (Figure 5B). When clear associations between GVHD and expression variants are defined, functional assays may ultimately require assessment of individual alleles, genes, haplotypes, cellular compartments, and tissues.97-99
Typing for SNPs. (A) TaqMan assay for typing biallelic SNPs. Actual TaqMan typing result for the G→A rs2075800 SNP (E→K at residue 602) of HSPA1L, a marker for GVHD in HLA-mismatched unrelated donor HCT.41 The assay is a real-time polymerase chain reaction (PCR) method in which the wild-type allele is PCR-amplified separately from the alternative allele using fluorescent allele-specific probes. Shown is the allelic discrimination plot in reverse orientation: CC homozygous (blue circles; reverse orientation for GG); TT homozygous (red circles; reverse orientation for AA); and CT heterozygous (green circles). Fluorescent signals are interpreted with the use of software that detects the presence of one dye (either homozygous wild-type represented by 1 color or homozygous alternative allele represented by a different color) or 2 dyes (heterozygous wild-type and alternative allele). (B) Sequence-based typing of SNPs. Actual sequence chromatogram (reverse orientation) of a MICA*008 018-heterozygous sample illustrating the C (blue)→T (red) substitution corresponding to valine→methionine at residue 129 (*). In this sample, MICA*008 encodes valine and MICA*018 encodes methionine. Methods are previously described by Moran et al.96
Typing for SNPs. (A) TaqMan assay for typing biallelic SNPs. Actual TaqMan typing result for the G→A rs2075800 SNP (E→K at residue 602) of HSPA1L, a marker for GVHD in HLA-mismatched unrelated donor HCT.41 The assay is a real-time polymerase chain reaction (PCR) method in which the wild-type allele is PCR-amplified separately from the alternative allele using fluorescent allele-specific probes. Shown is the allelic discrimination plot in reverse orientation: CC homozygous (blue circles; reverse orientation for GG); TT homozygous (red circles; reverse orientation for AA); and CT heterozygous (green circles). Fluorescent signals are interpreted with the use of software that detects the presence of one dye (either homozygous wild-type represented by 1 color or homozygous alternative allele represented by a different color) or 2 dyes (heterozygous wild-type and alternative allele). (B) Sequence-based typing of SNPs. Actual sequence chromatogram (reverse orientation) of a MICA*008 018-heterozygous sample illustrating the C (blue)→T (red) substitution corresponding to valine→methionine at residue 129 (*). In this sample, MICA*008 encodes valine and MICA*018 encodes methionine. Methods are previously described by Moran et al.96
Future needs to facilitate the translation of association-mapping data for clinical purposes will require careful assessment of the relative importance of genetic and nongenetic factors to outcome. In allogeneic transplantation, where disease diagnosis and stage, patient/donor age, sex, ABO blood type, and cytomegalovirus serostatus may all be important risk factors,100 the relative importance of each of these variables to genetic factors remains an important research objective. This information will facilitate the prospective selection of potential transplant donors, especially when patients have a choice among several donors.
Conclusions
The MHC remains a model system for understanding the immunogenetic basis of GVHD and transplant outcomes. GVHD is a polygenic disease where risk is contributed by the MHC haplotypes of the patient and the transplant donor. A new paradigm is emerging that includes consideration for both HLA coding and HLA haplotype–linked variation as important factors in unrelated donor HCT. The identification of specific novel variants within the MHC that play a role in GVHD underscores the need for more complete information on MHC haplotype diversity and the organization of sequence variation in ethnically diverse populations.
Acknowledgments
The author thanks Dr Mari Malkki and Dr Tao Wang for assistance with the figures, and Ms Courtney Preusse for preparation of the manuscript.
This study was supported by the National Institutes of Health, National Institute of Allergy and Infectious Diseases (AI069197) and the National Cancer Institute (CA100019, CA18029).
Authorship
Contribution: E.W.P. wrote the manuscript.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Effie W. Petersdorf, Fred Hutchinson Cancer Research Center, Division of Clinical Research, D4-115, 1100 Fairview Ave North, Seattle, WA 98109; e-mail: epetersd@fhcrc.org.