Abstract
The PIT1/SLC20A1 protein, a well-described sodium/phosphate cotransporter and retrovirus receptor, has been identified recently as a modular of proliferation and apoptosis in vitro. The targeted deletion of the PIT1 gene in mice revealed a lethal phenotype due to severe anemia attributed to defects in liver development. However, the presence of immature erythroid cells associated with impaired maturation of the globin switch led us to investigate the role of PIT1 in hematopoietic development. In the present study, specific deletion of PIT1 in the hematopoietic system and fetal liver transplantation experiments demonstrated that anemia was associated with an erythroid cell– autonomous defect. Moreover, anemia was not due to RBC destruction but rather to maturation defects. Because Erythroid Krüppel-like Factor (EKLF)–knockout mice showed similar maturation defects, we investigated the functional link between PIT1 and EKLF. We demonstrated that EKLF increases PIT1 expression during RBC maturation by binding to its promoter in vivo and that shRNA-driven depletion of either PIT1 or EKLF impairs erythroid maturation of G1E cells in vitro, whereas reexpression of PIT1 in EKLF-depleted G1E cells partially restores erythroid maturation. This is the first demonstration of a physiologic involvement of PIT1 in erythroid maturation in vivo.
Key Points
EKLF regulates PiT1 expression during erythroid maturation.
PiT1 is mandatory for erythroid maturation.
Introduction
PIT1/SLC20A1 initially identified as retrovirus receptors1,2 is a plasma membrane protein belonging to the SLC20A family of mammalian Na+-dependent phosphate (Pi) transporters.3,4 The broad tissue distribution of PIT1 first led to the assumption that it played an unregulated housekeeping role in supplying Pi to the cells.4,5 However, deletion of the PIT1 gene in mice revealed an unexpected phenotype. PIT1-knockout (KO) mice (PIT1Δ5/Δ5) died in utero at embryonic day 12.5 (E12.5) from severe anemia probably resulting from a defect in liver development.6 Moreover, mice carrying 2 copies of hypomorphic PIT1 alleles (PIT1neo/neo) expressing only 15% of wild-type PIT1 mRNA survived at birth but were growth retarded and anemic. The combination of both hypomorphic and null alleles (PIT1neo/Δ5), resulting in 6% of PIT1 expression, led to a late embryonic lethality (E15.5) with an intermediate phenotype between null and hypomorphic mice.6 Although most of the phenotypic features displayed by PIT1 mutant mice were attributed to a liver defect, analysis of peripheral blood smears from PIT1Δ5/Δ5 animals revealed a higher proportion of large nucleated erythroid cells associated with a higher ratio between fetal and adult forms of hemoglobin compared with their wild-type counterparts,6 suggesting a possible defect in erythroid cell maturation.
It is now well documented that during hematopoiesis, the expression patterns of specific transcription factors are crucial for the establishment, maturation, and maintenance of the different hematopoietic lineages.7,8 The Erythroid Krüppel-like Factor (EKLF/KLF1), the founding member of the KLF family of transcription factors, plays a crucial role in the development of RBCs.
EKLF is a DNA-binding protein defined by the presence of 3 highly similar C2H2-type zinc fingers at the C-terminus.9 It is implicated in β-like globin switching and adult β-globin gene regulation10,11 and is essential for the regulation of numerous erythroid genes, including components of the RBC membrane and cytoskeleton,12-14 heme synthesis enzymes,13 globin stabilizing protein (AHSP),12,13 and cell-cycle regulator proteins.15-18 Accordingly, EKLF has been implicated in hemoglobinization, cell-cycle arrest, and terminal differentiation of erythrocytes in vitro.19 EKLF levels influence the bipotential lineage decision directly by repressing megakaryocytic genes and stimulating erythroid genes.20-23 During hematopoiesis, EKLF is first expressed at low levels in common myeloid progenitors and becomes highly expressed only in megakaryocyte-erythroid bipotential progenitors while remaining elevated selectively in the terminally differentiated RBC progeny. The physiologic role of EKLF has been illustrated through the phenotypical analysis of EKLF-KO mice, which die in utero at E14.5 from severe anemia resulting from a failure of erythroid terminal differentiation.15,24,25 The phenotypic features of PIT1 mutant animals, similar to those of EKLF-KO mice, prompted us to investigate the functional interaction between EKLF and PIT1 during erythropoiesis.
The results of the present study demonstrate that the anemia observed in PIT1 mutant mice arises from a cell-autonomous defect in RBC maturation in vivo. We illustrate an EKLF-driven up-regulation of PIT1 during erythroid maturation in vivo. Accordingly, an underexpression of PIT1 in vitro led to an impaired differentiation of primary erythroid progenitors, recapitulating the erythroid maturation defect of EKLF-depleted cells. The abnormal maturation of EKLF-depleted erythroid cells could be partially rescued by PIT1 overexpression. These results show that PIT1 is a critical factor for normal erythroid maturation in vivo.
Methods
Animals
All mouse models used in the study were on a 129sv/J × C57BL/6J mixed background. Animal care and maintenance were provided through the University Paris Descartes accredited animal facility at Necker Faculty of Medicine (Paris). Mice were maintained on standard rodent laboratory chow (Special Diet Services). All procedures were approved by the Animal Care and Use Committee of the University Paris Descartes. PIT1 genetically modified mice were generated as described previously.6 Mx1-Cre;PIT1lox/lox mice were obtained by crossing Mx1-Cre mice26 with PIT1lox/lox mice. Tissue-specific deletion of PIT1 was induced by polyinosinic-polycytidylic acid (pIpC) injection as described previously,26 at 5 weeks of age.
Erythrocyte turnover analysis
Erythrocytes were biotinylated by tail vein injection of 3 mg of EZ-Link Sulfo-NHS-biotin (Pierce) dissolved in 0.2 mL of PBS. Four hours after injection, 3 μL of blood was drawn from the tail vein to determine the 100% biotinylated erythrocyte starting point. Subsequently, 3 μL of blood was drawn weekly for turnover determination. Erythrocytes were then resuspended in PBS, labeled with phycoerythrin-conjugated streptavidin (Molecular Probes), and analyzed by flow cytometry.
Hematopoietic progenitor cell assays
Single-cell suspensions from adult spleens were plated in 3 mL of Methocult (STEMCELL Technologies) in 35-mm bacterial Petri dishes and incubated at 37°C in a humidified atmosphere containing 5% CO2 in air. Colonies were scored at day 4 for CFU-granulocyte macrophage (CFU-GM) and CFU-erythroid (CFU-E) cells.
2,3-DPG measurement
Blood of mice was drawn from the tail vein and 2,3-diphosphoglycerate (2,3-DPG) was measured using the UV test (Roche) according to the manufacturer's instructions.
Benzidine staining
A total of 45 μL of cell suspension was incubated with 5 μL of 2% benzidine solution (Sigma-Aldrich) containing 2% H2O2 and counted immediately for blue staining under an optical microscope.
Transplantation experiments
E12 fetal livers from 3 PIT1Δ5/Δ5 embryos were pooled, mechanically dissociated by passing through a 23-G needle, and then filtered through a 70-μm cell strainer to obtain single-cell suspensions. RBCs were eliminated by a brief incubation in 0.8% ammonium chloride and the cell suspension was numerated for viable nucleated cells. Viable fetal liver nucleated cells were also obtained from livers of PIT1+/+ littermates. Sublethally irradiated (15Gy) Rag-KO (Ly5.1) mice were then retroorbitally injected with 1-3 × 105 viable fetal liver cells from E12 PIT1Δ5/Δ5 or PIT1+/+ embryos (Ly5.2).
Flow cytometric analysis
E12-E14.5 fetal livers, as well as BM and spleens from adult mice, were passed through a 23-G needle and filtered through a 70-μm cell strainer to obtain single-cell suspensions. Cells were incubated with conjugated Abs for 30 minutes at 4°C, briefly washed in PBS-5% BSA, and analyzed using a FACSCalibur or FORTESSA flow cytometer (BD Biosciences) after propidium iodide or SYTOX blue (Invitrogen) staining, allowing for the exclusion of dead cells. Abs used are listed in Table 1. Bromodeoxyuridine (BrdU) and annexin V staining were performed with the BrdU flow kit and the Annexin V apoptosis detection kit (BD Biosciences), respectively, according to the manufacturer's instructions.
Mouse PIT1 promoter cloning in pGL3 vector
A genomic DNA fragment containing 5000 bp of mouse PIT1 5′-flanking sequence was cloned upstream of a luciferase reporter gene in a promoterless expression vector (pGL3-Basic; Promega). Cloning of this 5-kb fragment was performed by homologous recombination in Escherichia coli using a recombineering approach.27 We used a PIT1 gene-containing BAC (RP23-160G19 clone; Invitrogen) as a template and conveniently modified primers (Table 2).
Cloning and mutagenesis primers
| Application . | Gene . | . | 5′-3′ sequence . |
|---|---|---|---|
| Cloning | Distal mouse PIT1 promoter | Forward | CAACGCGTGCGGCCATCATATAGAGAGCA |
| Reverse | AGAATTCCAAGGGCAAGTCTCAGCACAC | ||
| Cloning | Proximal PIT1 promoter | Forward | AGAATTCGGGACATGGGAACACTCACTG |
| Reverse | CATAGATCTCAGCAACTCCAAGGCAAGGT | ||
| Mutagenesis | EKLF1 | Forward | GTGTTTGGATATGCTAGTTTAGGGGAGCCTTACACG |
| Reverse | CGTGTAAGGCTCCCCTAAACTAGCATATCCAAACAC | ||
| Mutagenesis | EKLF2 | Forward | CTGGGAACGCAAGCCTATCC |
| Reverse | TACCTCGCCCATCTCACGTG | ||
| Mutagenesis | EKLF2 | Forward | CACGTGAGATGGGCGAGGTA |
| Reverse | GGGCGCGCCTAGCCG |
| Application . | Gene . | . | 5′-3′ sequence . |
|---|---|---|---|
| Cloning | Distal mouse PIT1 promoter | Forward | CAACGCGTGCGGCCATCATATAGAGAGCA |
| Reverse | AGAATTCCAAGGGCAAGTCTCAGCACAC | ||
| Cloning | Proximal PIT1 promoter | Forward | AGAATTCGGGACATGGGAACACTCACTG |
| Reverse | CATAGATCTCAGCAACTCCAAGGCAAGGT | ||
| Mutagenesis | EKLF1 | Forward | GTGTTTGGATATGCTAGTTTAGGGGAGCCTTACACG |
| Reverse | CGTGTAAGGCTCCCCTAAACTAGCATATCCAAACAC | ||
| Mutagenesis | EKLF2 | Forward | CTGGGAACGCAAGCCTATCC |
| Reverse | TACCTCGCCCATCTCACGTG | ||
| Mutagenesis | EKLF2 | Forward | CACGTGAGATGGGCGAGGTA |
| Reverse | GGGCGCGCCTAGCCG |
Mouse PIT1 promoter mutant constructs
EKLF putative consensus binding sites were modified to generate PIT1 promoter-containing EKLF binding site mutants (ΔEKLF1 and ΔEKLF2). ΔEKLF1 was obtained by deleting the 4-bp core sequence of the putative distal consensus binding site using the QuickChange II XL site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. ΔEKLF2 was generated by deleting the proximal EKLF putative consensus binding site using PCR overlap extension, as described previously.28 Using the same primers, double mutants (ΔEKLF1/ΔEKLF2) were obtained by PCR overlap extension using the ΔEKLF1 mutant as a template. The sequences of primers used are listed in Table 2. Shorter fragments of the mouse PIT1 promoter were obtained by digesting the 5000-bp promoter cloned into the pGL3-Basic vector using MluI/AflII (−1189/+1), MluI/PvuII (−543/+1), and MluI/BssHII (−119/+1), followed by a religation step.
Luciferase experiments
Twenty-four hours after seeding, cells were cotransfected with 1 μg of pGL3 reporter plasmid-containing PIT1 promoter constructs and 500 ng of various hematopoietic transcription factors expression plasmid (BioValley). To correct for transfection efficiency, all cells were cotransfected with 100 ng of the pRL-tk plasmid (Promega), expressing the Renilla reniformis luciferase. Negative controls were performed by cotransfecting cells either with a pGL3 plasmid containing the PIT1 promoter constructs and an empty expression vector or with an empty pGL3 plasmid and a hematopoietic transcription factor expression vector. The activities of firefly and Renilla luciferases were analyzed 48 hours after transfection using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer's instructions. The firefly luciferase activity was normalized for Renilla luciferase expression.
Chromatin immunoprecipitation (ChIP)
G1E cells were fixed with 0.75% formaldehyde in PBS for 10 minutes at room temperature and the reaction was quenched by the addition of glycine (125mM) for 5 minutes at room temperature. Cells were washed once in PBS and twice in immunoprecipitation buffer, resuspended in sonication buffer solution, and passed through a 23-G needle. Buffer compositions are given in Table 3. The chromatin was fragmented to approximately 300- to 1000-bp fragments by sonication, and then 25 μg of sonicated DNA was incubated overnight at 4°C with 4 μg of anti-EKLF Ab (ab2483; Abcam). A negative control was performed by incubating sonicated DNA without EKLF Ab. Protein A beads were incubated with 100 ng/μL of herring sperm DNA overnight at 4°C. The following day, 20 μL of saturated protein A beads per microgram of EKLF Ab was added to the sonicated DNA and incubated for 1 hour at 4°C. Five consecutive washes were then performed using salt buffer and Tris-EDTA buffer. Beads were eluted in 10% Chelex (Bio-Rad) and boiled for 10 minutes. Proteins were digested by incubation with 0.1 mg/mL of proteinase K for 30 minutes at 55°C. The reaction was stopped at 100°C for 10 minutes. Immunoprecipitated DNA was then analyzed by real-time PCR. Primers were designated to amplify 100-bp amplicons distributed along the PIT1 mouse 5000-bp promoters (Table 4). A positive control was performed using input DNA as a template.
Buffer compositions
| Application . | Buffer . | Composition . |
|---|---|---|
| ChIP | IP buffer | 150mM NaCl, 50mM Tris, 5mM EDTA, 0.5% NP-40, 1% Triton X-100, and 1× protease inhibitor cocktail from Roche |
| ChIP | Sonication buffer | 140mM NaCl, 50mM HEPES, 1mM EDTA, 1% Triton X-100, 0.1% NaDOC, 0.1% SDS, and 1× protease inhibitor cocktail |
| ChIP | Wash buffer I | 1% Triton, 0.1% NaDOC, 150mM NaCl, and 10mM Tris-Cl, pH 8 |
| ChIP | Wash buffer II | 1% NP-40, 1% NaDOC, 150mM NaCl, and 10mM Tris-Cl |
| ChIP | Wash buffer III | 0.5% Triton, 0.1% NaDOC, 500mM NaCl, and 10mM Tris-Cl |
| ChIP | Wash buffer IV | 0.5% NP-40, 0.5% NaDOC, 250mM LiCl, 20mM Tris-Cl, and 1mM EDTA |
| ChIP | Wash buffer V | 0.1% NP-40, 150mM NaCl, 20mM Tris-Cl, and 1mM EDTA |
| ChIP | TE | 10mM Tris-Cl, pH 8, and 1mM EDTA |
| Western blotting | Lysis buffer | 150mM NaCl, 10mM Tris HCl, 5mM EDTA, 1% NP-40, 0.1% SDS, 0.5% DOC, 1mM Na3VO4, 1mM NaF, 5mM Na pyrophosphate, and a protease inhibitor cocktail |
| Western blotting | TBST | 10mM Tris, pH 7.5, 154mM NaCl, and 0.15% Tween 20 |
| Application . | Buffer . | Composition . |
|---|---|---|
| ChIP | IP buffer | 150mM NaCl, 50mM Tris, 5mM EDTA, 0.5% NP-40, 1% Triton X-100, and 1× protease inhibitor cocktail from Roche |
| ChIP | Sonication buffer | 140mM NaCl, 50mM HEPES, 1mM EDTA, 1% Triton X-100, 0.1% NaDOC, 0.1% SDS, and 1× protease inhibitor cocktail |
| ChIP | Wash buffer I | 1% Triton, 0.1% NaDOC, 150mM NaCl, and 10mM Tris-Cl, pH 8 |
| ChIP | Wash buffer II | 1% NP-40, 1% NaDOC, 150mM NaCl, and 10mM Tris-Cl |
| ChIP | Wash buffer III | 0.5% Triton, 0.1% NaDOC, 500mM NaCl, and 10mM Tris-Cl |
| ChIP | Wash buffer IV | 0.5% NP-40, 0.5% NaDOC, 250mM LiCl, 20mM Tris-Cl, and 1mM EDTA |
| ChIP | Wash buffer V | 0.1% NP-40, 150mM NaCl, 20mM Tris-Cl, and 1mM EDTA |
| ChIP | TE | 10mM Tris-Cl, pH 8, and 1mM EDTA |
| Western blotting | Lysis buffer | 150mM NaCl, 10mM Tris HCl, 5mM EDTA, 1% NP-40, 0.1% SDS, 0.5% DOC, 1mM Na3VO4, 1mM NaF, 5mM Na pyrophosphate, and a protease inhibitor cocktail |
| Western blotting | TBST | 10mM Tris, pH 7.5, 154mM NaCl, and 0.15% Tween 20 |
DOC indicates deoxycholate.
PCR primers
| . | . | 5′-3′ sequence . |
|---|---|---|
| Mouse PIT1 promoter | Forward | TACATGGGGAAAGGGAAAGGAC |
| Reverse | GGGGACATGGGAACACTCACT | |
| Forward | GGTGGCTGGAGAAAGGAAGG | |
| Reverse | CAGGGCTGTTGGAAGGAGTAAA | |
| Forward | CACACCAGAAGAGGGCATCG | |
| Reverse | CCTCCCCCATCAATCCCTAAT | |
| Forward | GACTGAAGCAAAGCACATCCAA | |
| Reverse | TTGCCCTTACTTCTTTCCAAGG | |
| Forward | CAAGGGCAAGTCTCAGCACAC | |
| Reverse | GGGGACTGTGGGGGAGTCT | |
| Mouse PIT1 | Forward | CTTCCTTGTTCGTGCGTTCAT |
| Reverse | AAGAGGTTGATTCCGATTGTGC | |
| Mouse Pinin | Forward | ATTGCTGGCCCTTTCTGGT |
| Reverse | GGGGGTCCTCCTCCTCCACTA | |
| Mouse EKLF | Forward | TTCCTCAAGTGGTGGCGGT |
| Reverse | TCCTCCGATTTCAGACTCACG | |
| Mouse β-globin | Forward | GGCAGGCTGCTGGTTGTCTA |
| Reverse | GCCATGGGCCTTCACTTTG | |
| Mouse P21 | Forward | GAAAACGGAGGCAGACCAGC |
| Reverse | CCTCCTGACCCACAGCAGAA | |
| Mouse Erythropoietin | Forward | GTCCCACCCTGCTGCTTTTAC |
| Reverse | CCAGAACTCGACTGTCGCAGA | |
| Mouse Dematin | Forward | CCAACCAGCCAGCCAAGATA |
| Reverse | TTCGAGATGCCTTCCGTTTC |
| . | . | 5′-3′ sequence . |
|---|---|---|
| Mouse PIT1 promoter | Forward | TACATGGGGAAAGGGAAAGGAC |
| Reverse | GGGGACATGGGAACACTCACT | |
| Forward | GGTGGCTGGAGAAAGGAAGG | |
| Reverse | CAGGGCTGTTGGAAGGAGTAAA | |
| Forward | CACACCAGAAGAGGGCATCG | |
| Reverse | CCTCCCCCATCAATCCCTAAT | |
| Forward | GACTGAAGCAAAGCACATCCAA | |
| Reverse | TTGCCCTTACTTCTTTCCAAGG | |
| Forward | CAAGGGCAAGTCTCAGCACAC | |
| Reverse | GGGGACTGTGGGGGAGTCT | |
| Mouse PIT1 | Forward | CTTCCTTGTTCGTGCGTTCAT |
| Reverse | AAGAGGTTGATTCCGATTGTGC | |
| Mouse Pinin | Forward | ATTGCTGGCCCTTTCTGGT |
| Reverse | GGGGGTCCTCCTCCTCCACTA | |
| Mouse EKLF | Forward | TTCCTCAAGTGGTGGCGGT |
| Reverse | TCCTCCGATTTCAGACTCACG | |
| Mouse β-globin | Forward | GGCAGGCTGCTGGTTGTCTA |
| Reverse | GCCATGGGCCTTCACTTTG | |
| Mouse P21 | Forward | GAAAACGGAGGCAGACCAGC |
| Reverse | CCTCCTGACCCACAGCAGAA | |
| Mouse Erythropoietin | Forward | GTCCCACCCTGCTGCTTTTAC |
| Reverse | CCAGAACTCGACTGTCGCAGA | |
| Mouse Dematin | Forward | CCAACCAGCCAGCCAAGATA |
| Reverse | TTCGAGATGCCTTCCGTTTC |
Cell culture conditions and transfections/infections
Primary erythroid progenitors isolated from fetal livers of PIT1+/+ and PIT1neo/Δ5 fetuses and from the BM and spleens of PIT1+/+ and PIT1neo/neo adult mice were cultured in proliferation medium for 3-7 days until 90% purity of erythroid cells was obtained, and then in differentiation medium as described previously.29 HEK cells were maintained in DMEM/nutrient mixture F12 supplemented with 0.1% bicarbonate, 2mM glutamine, 12mM HEPES, and 10% FBS. G1E-ER-Gata1 cells were cultured in IMDM supplemented with 15% FBS, 4.5 × 10−5M monothioglycerol (Sigma-Aldrich), 2 U/mL of erythropoietin (Epo), and 50 ng/mL of SCF. For plasmid transfection, HEK cells were seeded 24 hours before the experiment in antibiotic-free medium at 160 000 cells per well in a 24-well plate. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. G1E cells were infected with MISSION shRNA lentiviral transduction particles (Sigma-Aldrich) and human PIT1 inserted into pHAGE-CMV-MCS-IZsGreenW lentiviral vector30 in culture medium supplemented with 8 μg/mL of polybrene. Transduced cells were selected using puromycin (1.5 μg/mL) or cell sorting.
Gene expression
Total RNA was isolated from cells and tissues using NucleoSpin RNA II columns (Macherey Nagel). RT-PCR amplifications were performed using M-MLV (Invitrogen) according to the manufacturer's instructions. Real-time PCR was performed using SYBR Green chemistry (Thermo Scientific) on an ABI Prism 7000 detection system. The pinin gene was used as the reference gene31 and expression differences were calculated as described previously.32 Primers used are listed in Table 4.
Western blotting
Proteins were prepared and analyzed as described previously.30 The buffers used are listed in Table 2 and Abs in Table 5.
Western blot Abs
| Antigen . | Dilution . | Clone . |
|---|---|---|
| Human PIT1 | 1/1000 | Homemade41 |
| Mouse PIT1 | 1/1000 | Homemade with previously described protocol41 |
| Mouse EKLF | 1/500 | Ab38245 (Abcam) |
| β-Actin | 1/1000 | AC74 (Sigma-Aldrich) |
| β-Tubulin | 1/2500 | TUB2,1 (Sigma-Aldrich) |
Statistical analysis
Results are presented as means ± SEM. Statistical analyses were performed using either the Student t test or the alternate Welch t test when SDs were different or the nonparametric Mann-Whitney U test. In all cases, the level of statistical significance was set at P < .05.
Micrograph analysis
The digital format of slide images were obtained using NanoZoomer Digital Pathology 2.0HT virtual microscope (Hamamatsu Photonics) containing a high-speed and high-resolution digital-slide scanner. Whole slide images were generated using a 20× magnification. Micrographs were then analyzed using NDP view Version 1.2.23 software (Hamamatsu Photonics).
Results
Hypomorphic PIT1 expression in mice leads to hematopoietic stress
As described previously, the hypomorphic expression of PIT1 in adult mice led to a decrease in RBC count and hemoglobin concentration6 (Table 6). We determined the RBC half-life (Figure 1A) and found no significant difference between PIT1neo/neo mice (17.6 ± 1.5 days) and wild-type mice (20.1 ± 1.1 days), demonstrating that an increase in peripheral RBC destruction was not the cause of the observed anemia.
Hematologic variables in adult mice according to genotype
| . | PIT1+/+ . | PIT1neo/neo . | PIT1lox/lox . | Mx1-Cre-PIT1lox/lox . |
|---|---|---|---|---|
| RBCs, 106/mm3 | 11.1 ± 0.5 | 8.7 ± 1.2** | 9.1 ± 0.3 | 7.6 ± 0.2** |
| Hemoglobin, g/dL | 18.1 ± 1.2 | 14.6 ± 1.8** | 18.7 ± 0.7 | 18.9 ± 0.7 |
| Mean corpuscular hemoglobin, pg | 16.4 ± 0.6 | 16.8 ± 0.3 | 24.9 ± 0.9 | 20.6 ± 0.9* |
| Mean corpuscular volume, μm | 56.2 ± 1.3 | 59.8 ± 2.4* | 51.4 ± 2.4 | 55.7 ± 4.5 |
| WBCs, 103/mm3 | 9.3 ± 1.8 | 10.2 ± 5.2 | 6.2 ± 1.9 | 9.1 ± 1.1* |
| Platelets, 103/mm3 | 676 ± 271 | 814 ± 462 | 461 ± 91 | 331 ± 101 |
| Reticulocytes, % | 5.2 ± 0.8 | 13.2 ± 1.4** | 2.3 ± 0.3 | 4.1 ± 0.3* |
| . | PIT1+/+ . | PIT1neo/neo . | PIT1lox/lox . | Mx1-Cre-PIT1lox/lox . |
|---|---|---|---|---|
| RBCs, 106/mm3 | 11.1 ± 0.5 | 8.7 ± 1.2** | 9.1 ± 0.3 | 7.6 ± 0.2** |
| Hemoglobin, g/dL | 18.1 ± 1.2 | 14.6 ± 1.8** | 18.7 ± 0.7 | 18.9 ± 0.7 |
| Mean corpuscular hemoglobin, pg | 16.4 ± 0.6 | 16.8 ± 0.3 | 24.9 ± 0.9 | 20.6 ± 0.9* |
| Mean corpuscular volume, μm | 56.2 ± 1.3 | 59.8 ± 2.4* | 51.4 ± 2.4 | 55.7 ± 4.5 |
| WBCs, 103/mm3 | 9.3 ± 1.8 | 10.2 ± 5.2 | 6.2 ± 1.9 | 9.1 ± 1.1* |
| Platelets, 103/mm3 | 676 ± 271 | 814 ± 462 | 461 ± 91 | 331 ± 101 |
| Reticulocytes, % | 5.2 ± 0.8 | 13.2 ± 1.4** | 2.3 ± 0.3 | 4.1 ± 0.3* |
The circulating blood of 3-month-old PIT1+/+, PIT1neo/neo, PIT1lox/lox, and Mx1-Cre;PIT1lox/lox mice was analyzed on a Vet ABC counter (SCIL). The data shown were obtained from a group of 5 mice. Values are means ± SEM.
P < .05 and **P < .01 for significant differences between PIT1neo/neo and PIT1+/+ mice and between Mx1-Cre;PIT1lox/lox mice and PIT1lox/lox controls.
PIT1 deletion results to RBC defects in adult mice. (A) Turnover analysis of RBCs in adult mice constitutively underexpressing PIT1 (PIT1neo/neo). Erythrocytes were biotinylated by tail vein injection and blood was drawn weekly. RBCs of PIT1+/+ (black diamonds) and PIT1neo/neo (white squares) mice were labeled with PE-conjugated streptavidin and analyzed by flow cytometry. Erythrocyte survival was determined by the assessment of the number of biotinylated erythrocytes relative to the starting level (100% biotinylated). (B) Six-week-old PIT1+/+ mice spleen (left) and PIT1neo/neo mice spleen (right). (C) Spleen section stained with H&E at 2.5 and 40× magnification. (D) In vitro differentiation of adult hematopoietic spleen cells. The number of CFU-GM and CFU-E per 105 nucleated spleen cells are indicated. (E) Representative flow cytometric analysis of PIT1+/+ and PIT1neo/neo adult spleen. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (G) Measurement of 2,3-DPG content of RBCs. Data indicated means ± SEM of at least 3 animals per condition. Significant differences from PIT1+/+ mice (black bar) are indicated. *P < .05; ***P < .001).
PIT1 deletion results to RBC defects in adult mice. (A) Turnover analysis of RBCs in adult mice constitutively underexpressing PIT1 (PIT1neo/neo). Erythrocytes were biotinylated by tail vein injection and blood was drawn weekly. RBCs of PIT1+/+ (black diamonds) and PIT1neo/neo (white squares) mice were labeled with PE-conjugated streptavidin and analyzed by flow cytometry. Erythrocyte survival was determined by the assessment of the number of biotinylated erythrocytes relative to the starting level (100% biotinylated). (B) Six-week-old PIT1+/+ mice spleen (left) and PIT1neo/neo mice spleen (right). (C) Spleen section stained with H&E at 2.5 and 40× magnification. (D) In vitro differentiation of adult hematopoietic spleen cells. The number of CFU-GM and CFU-E per 105 nucleated spleen cells are indicated. (E) Representative flow cytometric analysis of PIT1+/+ and PIT1neo/neo adult spleen. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (G) Measurement of 2,3-DPG content of RBCs. Data indicated means ± SEM of at least 3 animals per condition. Significant differences from PIT1+/+ mice (black bar) are indicated. *P < .05; ***P < .001).
Because the spleens of PIT1neo/neo mice were enlarged, redder (Figure 1B), and histologically disorganized (Figure 1C), we conducted in vitro hematopoietic progenitor assays (Figure 1D). Results revealed an increased number of GM and erythroid numbers compared with PIT1+/+ animals. Moreover, cell surface marker analysis by FACS showed and increased number of immature RBCs (Figure 1E). These results suggest that hematopoietic stress occurs in the spleens of PIT1neo/neo mice, probably to compensate for anemia, and that maturation of their RBCs could be impaired.
In response to anemia, it is known that the 2,3-DPG content of RBCs is increased to improve oxygen release to tissues. Unexpectedly, 2,3-DPG levels were decreased by 27% in PIT1neo/neo mice (Figure 1F), suggesting an impairment of RBCs to respond to anemia. The decrease in 2,3-DPG level observed in PIT1neo/neo adult mice suggests that PIT1, functioning as a phosphate transporter, may play an important role in RBC metabolism for 2,3-DPG synthesis.
Underexpression of PIT1 specifically in the hematopoietic system at the adult stage recapitulates the hematopoietic phenotype of hypomorphic PIT1neo/neo mice
To better characterize the role of PIT1 in the onset of the observed anemia, we deleted the PIT1 gene specifically in the hematopoietic system. To do this, we crossed PIT1lox/lox mice to Mx1-Cre transgenic mice expressing the Cre recombinase transgene under the control of the Mx1-inducible promoter.26 Treatment of Mx1-Cre;PIT1lox/lox mice with pIpC at the adult stage led to a 63% decrease of PIT1 mRNA in BM cells 6 months after treatment (Figure 2A). Similar to that shown in PIT1neo/neo mice, peripheral blood analysis of Mx1-Cre;PIT1lox/lox mice revealed a decrease in RBC count compared with PIT1lox/lox mice (Table 6; supplemental Figure 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article). Moreover, spleens were enlarged (Figure 2B) and histology showed increased cellularity and disorganized red and white pulp (Figure 2C). In vitro hematopoietic progenitor assays from the spleens of Mx1-Cre;PIT1lox/lox animals (Figure 2D) showed an increase in GM and erythroid colony number and FACS analysis (Figure 2E) revealed an accumulation of immature RBCs, suggesting an impairment in their maturation. Measurement of 2,3-DPG levels indicated that it was decreased by 26% in RBCs from Mx1-Cre;PIT1lox/lox mice (Figure 2F).
Specific deletion of PIT1 at the adult stage results in hematopoietic defect. (A) Quantitative analysis of PIT1 mRNA expression in Mx1-Cre;PIT1lox/lox BM mice 6 months after pIpC treatment. (B) Six-month-old PIT1lox/lox mice spleen (top) and PIT1neo/neo mice spleen (bottom). (C) Spleen section stained with H&E at 2.5 and 40× magnification. (D) In vitro differentiation of adult hematopoietic spleen cells from Mx1-Cre;PIT1lox/lox. The numbers of CFU-GM and CFU-E per 105 nucleated spleen cells are indicated. (E) Representative flow cytometric analysis of PIT1lox/lox and Mx1-Cre;PIT1lox/lox adult spleen. Percentages of labeled cells were calculated by taking into account the background labeling (baseline define by omitting Ab). (F) Measurement of 2,3-DPG Mx1-Cre-PIT1lox/lox mice blood. Data indicate the means ± SEM of at least 5 animals per condition. Significant differences from PIT1lox/lox mice (black bar) are indicated. *P < .05; **P < .01; ***P < .001.
Specific deletion of PIT1 at the adult stage results in hematopoietic defect. (A) Quantitative analysis of PIT1 mRNA expression in Mx1-Cre;PIT1lox/lox BM mice 6 months after pIpC treatment. (B) Six-month-old PIT1lox/lox mice spleen (top) and PIT1neo/neo mice spleen (bottom). (C) Spleen section stained with H&E at 2.5 and 40× magnification. (D) In vitro differentiation of adult hematopoietic spleen cells from Mx1-Cre;PIT1lox/lox. The numbers of CFU-GM and CFU-E per 105 nucleated spleen cells are indicated. (E) Representative flow cytometric analysis of PIT1lox/lox and Mx1-Cre;PIT1lox/lox adult spleen. Percentages of labeled cells were calculated by taking into account the background labeling (baseline define by omitting Ab). (F) Measurement of 2,3-DPG Mx1-Cre-PIT1lox/lox mice blood. Data indicate the means ± SEM of at least 5 animals per condition. Significant differences from PIT1lox/lox mice (black bar) are indicated. *P < .05; **P < .01; ***P < .001.
Our results show that the hematopoietic phenotype seen in hypomorphic PIT1neo/neo mice could be recapitulated by specifically deleting PIT1 in hematopoietic tissue at the adult stage, strongly suggesting that the phenotypic alterations are not because of adverse effects of PIT1 deletion in tissue of nonhematopoietic origin. It is possible that WBC development was also altered in our mice models, because we observed increased splenic GM colony numbers (Figures 1D and 2D) and increased WBC count in Mx1-Cre;PIT1lox/lox mice (Table 6). Nevertheless, we will focus on the observed anemia and thus on the RBC defect.
E12 PIT1Δ5/Δ5 fetal liver cells do not reconstitute lethally irradiated mice
The anemia observed in PIT1neo/neo mice occurs despite a normal liver development. We conducted PIT1Δ5/Δ5 fetal liver transplantations to determine whether hematopoietic perturbations observed in PIT1neo/neo mice were intrinsic to stem/progenitor cells rather than caused by microenvironmental liver defects. Irradiated Rag KO recipient mice (Ly5.1) received transplantations from E12 PIT1Δ5/Δ5 fetal livers (Ly5.2) that do not express PIT1. Results shows that all recipient mice injected with E12 PIT1Δ5/Δ5 fetal liver cells died 1-2 weeks after irradiation. Conversely, E12 fetal liver cells from wild-type animals could reconstitute irradiated recipient mice because Ly5.2 staining was detected by FACS analysis in their BM and spleen cells (supplemental Figure 2). These results suggest that PIT1Δ5/Δ5 hematopoietic cells were not able to reconstitute irradiated mice in spite of a normal microenvironment, and demonstrate that a null mutation of PIT1 intrinsically impaired hematopoietic cell development.
PIT1 deletion impairs fetal erythropoiesis in mice
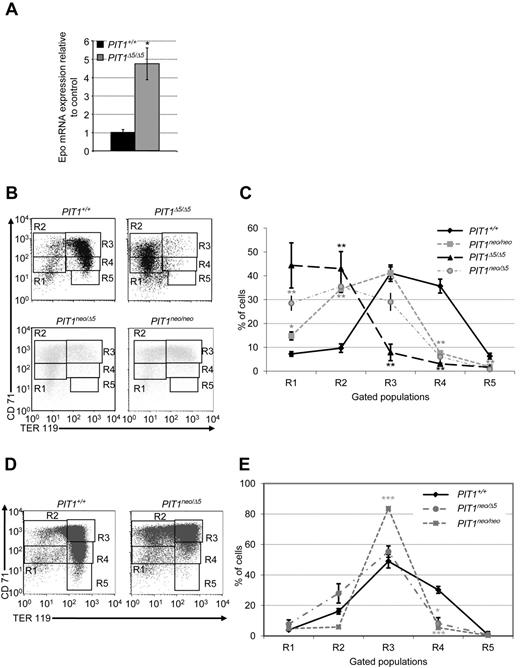
Because anemia is not likely to be caused by abnormal RBC destruction, we investigated whether it could be due to an intrinsic maturation defect. We first evaluated PIT1-depleted liver function by measuring Epo transcripts levels in E12 PIT1+/+ and PIT1Δ5/Δ5 fetal livers. Results demonstrated that the level of Epo transcripts was increased in mutant livers, suggesting a normal response of hepatoblasts to anemia (Figure 3A).
Disruption of PIT1 delays erythroid maturation in E12 and E14 fetal livers. (A) Quantification of Epo mRNAs in PIT1+/+ and PIT1Δ5/Δ5 E12 fetal livers by real-time PCR. Data indicate the means ± SEM of 5 animals per genotype. Significant differences from control (black bar) are indicated. *P < .05. (B,D) Representative flow cytometric analysis of E12 fetal liver cells from PIT1+/+, PIT1neo/neo, PIT1neo/Δ5, and PIT1Δ5/Δ5embryos (B) and E14.5 fetal liver cells from PIT1+/+, PIT1neo/Δ5 and PIT1neo/neo embryos (D). R1-R5 populations correspond to erythroid cells at different stages of maturation. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (C,E) Analysis of percentage of E12 (C) and E14.5 PIT1+/+ (black diamonds), PIT1neo/neo (gray squares), PIT1neo/Δ5 (gray circles) and PIT1Δ5/Δ5 (black triangles) fetal liver erythroid cells (E) in the different stages of maturation. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01; ***P < .001.
Disruption of PIT1 delays erythroid maturation in E12 and E14 fetal livers. (A) Quantification of Epo mRNAs in PIT1+/+ and PIT1Δ5/Δ5 E12 fetal livers by real-time PCR. Data indicate the means ± SEM of 5 animals per genotype. Significant differences from control (black bar) are indicated. *P < .05. (B,D) Representative flow cytometric analysis of E12 fetal liver cells from PIT1+/+, PIT1neo/neo, PIT1neo/Δ5, and PIT1Δ5/Δ5embryos (B) and E14.5 fetal liver cells from PIT1+/+, PIT1neo/Δ5 and PIT1neo/neo embryos (D). R1-R5 populations correspond to erythroid cells at different stages of maturation. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (C,E) Analysis of percentage of E12 (C) and E14.5 PIT1+/+ (black diamonds), PIT1neo/neo (gray squares), PIT1neo/Δ5 (gray circles) and PIT1Δ5/Δ5 (black triangles) fetal liver erythroid cells (E) in the different stages of maturation. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01; ***P < .001.
We next analyzed erythroid progenitor maturation by flow cytometry in PiTΔ5/Δ5 fetal livers at E12. At this stage, erythropoiesis is confined to the fetal liver, and discrete populations in particular stages of erythropoiesis can be separated using the cell-surface markers CD71 and TER119 (Figure 3B-C). Results showed that PIT1Δ5/Δ5 embryos contained early erythroid progenitors (R1) and erythroid progenitors (R2), whereas PIT1+/+ embryos mainly contained more mature populations (R3-R5).
The same experiments were repeated in PIT1neo/Δ5 E12 fetal liver cells, which express only 6% of PIT1.6 Results showed that 93% of erythroid cells were present in the R1-R3 population, with only 6% in R4 (Figure 3B-C). Interestingly, E12 PIT1neo/neo embryos, in which 15% of PIT1 was remaining, had an intermediate phenotype. These results suggest that PIT1 plays a role in erythroid maturation and that this is correlated with the level of PIT1 gene expression. Moreover, study of erythroid maturation in E14.5 PIT1neo/Δ5 embryos (Figure 3D-E) showed persistence in delayed maturation. In agreement with these observations, erythroid maturation observed in E14.5 PIT1neo/neo embryos was intermediate between PIT1neo/Δ5 and wild-type embryos.
The mouse PIT1 promoter is activated by EKLF in vitro
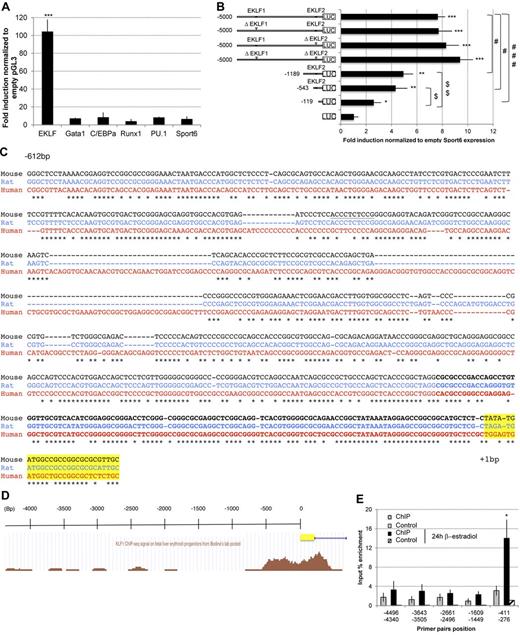
In silico analysis of mouse PIT1 promoter using Genomatix MatInspector Version 8.0 software revealed the presence of putative hematopoietic transcription factor consensus-binding sites. To investigate whether they could activate the PIT1 promoter using a luciferase assay system, a construct consisting of 5000 bp of the mouse PIT1 promoter cloned upstream of the firefly luciferase gene was cotransfected with different vectors expressing hematopoietic transcription factors along with a Renilla luciferase expression plasmid. Whereas Runx1, PU.1, C/EBPα, or Gata1 had no effect on luciferase activity alone (Figure 4A) or in combination (data not shown), EKLF was able to activate the 5000-bp mouse PIT1 promoter in vitro.
EKLF transactivates the mouse PIT1 promoter. (A) Luciferase assay on PIT1 5000-bp wild-type promoter. Results are expressed as the fold induction normalized with the empty pGL3-vector. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from Sport6 vector are indicated. ***P < .001. (B) Luciferase assay on PIT1 5000-bp wild-type promoter, EKLF binding site mutants, and wild-type promoter shorter constructs. Results are normalized with empty sport6 expression vector for each construct and are expressed as the fold induction. Data indicate the means ± SEM of 4 independent experiments. Significant differences from pGL3 (*P < .05; **P < .01; ***P < .001), from the mouse PIT1 wild-type promoter (#P < .05; ###P < .001), and from the −119-bp construct ($P < .05; $$P < .01) are indicated. (C) Phylogenetic alignment of the 600 proximal bp of the PIT1 promoter. Underlined sequence indicates EKLF2 consensus binding site, bold sequence indicates −119-bp construct, and highlighted sequence indicates the beginning of the PIT1 gene. Asterisks indicate nucleotide identities between the 3 species. (D) EKLF occupancy in the mouse PIT1 promoter using the UCSC browser as described by Bodin et al.33 (E) Quantitative PCR analysis of EKLF ChIP experiments. EKLF-immunoprecipitated DNA from G1E-ER-Gata1 cells treated or not with β-estradiol for 24 hours were analyzed by quantitative PCR using primers located along the mouse PIT1 promoter. Negative controls were performed using DNA incubated with beads but without anti-EKLF Ab. Means ± SEM of 4 experiments are presented. Significant differences from negative control are indicated. *P < .05.
EKLF transactivates the mouse PIT1 promoter. (A) Luciferase assay on PIT1 5000-bp wild-type promoter. Results are expressed as the fold induction normalized with the empty pGL3-vector. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from Sport6 vector are indicated. ***P < .001. (B) Luciferase assay on PIT1 5000-bp wild-type promoter, EKLF binding site mutants, and wild-type promoter shorter constructs. Results are normalized with empty sport6 expression vector for each construct and are expressed as the fold induction. Data indicate the means ± SEM of 4 independent experiments. Significant differences from pGL3 (*P < .05; **P < .01; ***P < .001), from the mouse PIT1 wild-type promoter (#P < .05; ###P < .001), and from the −119-bp construct ($P < .05; $$P < .01) are indicated. (C) Phylogenetic alignment of the 600 proximal bp of the PIT1 promoter. Underlined sequence indicates EKLF2 consensus binding site, bold sequence indicates −119-bp construct, and highlighted sequence indicates the beginning of the PIT1 gene. Asterisks indicate nucleotide identities between the 3 species. (D) EKLF occupancy in the mouse PIT1 promoter using the UCSC browser as described by Bodin et al.33 (E) Quantitative PCR analysis of EKLF ChIP experiments. EKLF-immunoprecipitated DNA from G1E-ER-Gata1 cells treated or not with β-estradiol for 24 hours were analyzed by quantitative PCR using primers located along the mouse PIT1 promoter. Negative controls were performed using DNA incubated with beads but without anti-EKLF Ab. Means ± SEM of 4 experiments are presented. Significant differences from negative control are indicated. *P < .05.
The mouse PIT1 promoter contains 2 putative EKLF consensus-binding sites located at position −3563 (EKLF1) and −450 (EKLF2). Deletion of the core sequence of these sites showed effects that are not consistent with the magnitude of PIT1 promoter activation by EKLF, suggesting that additional EKLF active sites may be present in the PIT1 promoter. Therefore, we tested a series of shorter PIT1 promoter constructs (Figure 4B). Results suggest that the region between −543 and −119 bp contains one or several critical EKLF-binding sites (Figure 4C), as previously reported from ChIP-seq experiments33 (Figure 4D).
EKLF interacts with the mouse PIT1 promoter in vivo
To determine whether the transcription factor EKLF could interact with the PIT1 promoter in vivo, we performed ChIP experiments in the mouse erythroid cell line G1E-ER-Gata1 cells34 treated or not with β-estradiol for 24 hours. These cells contained an Estrogen Receptor-Gata1 fusion protein, keeping Gata1 localized to the cytoplasm in the absence of ER ligand and thereby maintaining cells arrested in differentiation.35 These cells resemble primary erythroblasts, which, on addition of ER ligand, activates Gata1 target genes and initiates erythroid maturation.35
Immunoprecipitated DNA was analyzed by real-time PCR using primers located along the mouse PIT1 promoter (Figure 4E). Results showed DNA amplification with primers located at −411/−276 bp when G1E were treated with β-estradiol for 24 hours, but not with −4496/−4340 bp, −3643/−3505 bp, −2661/−2496 bp, or −1609/−1449 bp couples of primers, suggesting a specific binding of endogenous EKLF with the proximal region of the mouse PIT1 promoter in vivo during erythroid maturation. These results show endogenous interaction of EKLF with the PIT1 proximal promoter in a differentiating mouse erythroid cell line.
Knockdown of PIT1 impaired maturation of ex vivo cultured primary erythroid progenitors
To test the hypothesis of whether PIT1 could be involved in erythroid maturation, we cultured and differentiated primary erythroid progenitors isolated from BM of PIT1+/+ and PIT1neo/neo mice. PIT1 (Figure 5A) and EKLF (Figure 5B) gene expression were measured during maturation. Results showed that PIT1 expression increased over time and peaked 3 days after maturation induction in PIT1+/+ cells. Interestingly, EKLF expression peaked earlier (at day 2), which is consistent with the notion that EKLF could activate PIT1 expression during erythroid maturation. The increase of PIT1 expression was lower in PIT1neo/neo cells, which is consistent with hypomorphic allele expression, whereas the variation of EKLF expression was similar to that of wild-type mice. Cell maturation was monitored using benzidine staining (Figure 5C) and β-globin gene expression (Figure 5D). Benzidine staining showed increased hemoglobinization during differentiation for both genotypes, but to a lesser extent in PIT1neo/neo cells at day 2 and 3. This was confirmed by a lower β-globin gene expression at day 2 in PIT1neo/neo cells. We also analyzed cell-surface markers and found persistence of c-kit+ cells in erythroid progenitors culture from PIT1neo/neo mice (Figure 5E). These c-kit+ cells presented with a high forward scatter that was correlated with immature erythroid cells (Figure 5F; supplemental Figure 3). It is well documented that erythroid terminal differentiation needs proliferation arrest.16,18,19 EKLF-driven p21 increase has been reported to be necessary for this cell-cycle arrest.16 We showed (Figure 5G) that P21 gene expression strongly increased in PIT1+/+ cells at day 2, whereas this increase was lower in PIT1neo/neo cells, suggesting that PIT1neo/neo erythroid cells do not maturate properly because of defects in cell-cycle arrest. This was confirmed by cell-cycle analysis showing higher incorporation of BrdU at day 2 in CD71+ cells isolated from PIT1neo/neo mice, whereas PIT1+/+ cells accumulated in G1 phase of the cell cycle (supplemental Figure 4).
PIT1-underexpressing mice displayex vivoerythroid maturation defects. Primary erythroid progenitors were isolated from BM of PIT1+/+ and PIT1neo/neo mice and cultured ex vivo for 4 days. (A-B) PIT1 (A) and EKLF (B) mRNA expression in primary erythroid progenitors. Results are normalized to day 0 for each genotype. (C) Percentage of benzidine-positive cells during ex vivo maturation. (D) β-globin mRNA expression in PIT1neo/neo primary erythroid progenitors 2 days after ex vivo maturation induction compared with PIT1+/+ cells. (E) Percentage of c-kit+ primary erythroid progenitors determined by FACS analysis. (F) Representative flow cytometric analysis of primary erythroid progenitors 3 days after ex vivo maturation induction. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (G) p21 mRNA expression in primary erythroid progenitors during ex vivo maturation. Results are normalized to day 0 for each genotype. Data indicate the means ± SEM of at least 3 animals per genotype. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01.
PIT1-underexpressing mice displayex vivoerythroid maturation defects. Primary erythroid progenitors were isolated from BM of PIT1+/+ and PIT1neo/neo mice and cultured ex vivo for 4 days. (A-B) PIT1 (A) and EKLF (B) mRNA expression in primary erythroid progenitors. Results are normalized to day 0 for each genotype. (C) Percentage of benzidine-positive cells during ex vivo maturation. (D) β-globin mRNA expression in PIT1neo/neo primary erythroid progenitors 2 days after ex vivo maturation induction compared with PIT1+/+ cells. (E) Percentage of c-kit+ primary erythroid progenitors determined by FACS analysis. (F) Representative flow cytometric analysis of primary erythroid progenitors 3 days after ex vivo maturation induction. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (G) p21 mRNA expression in primary erythroid progenitors during ex vivo maturation. Results are normalized to day 0 for each genotype. Data indicate the means ± SEM of at least 3 animals per genotype. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01.
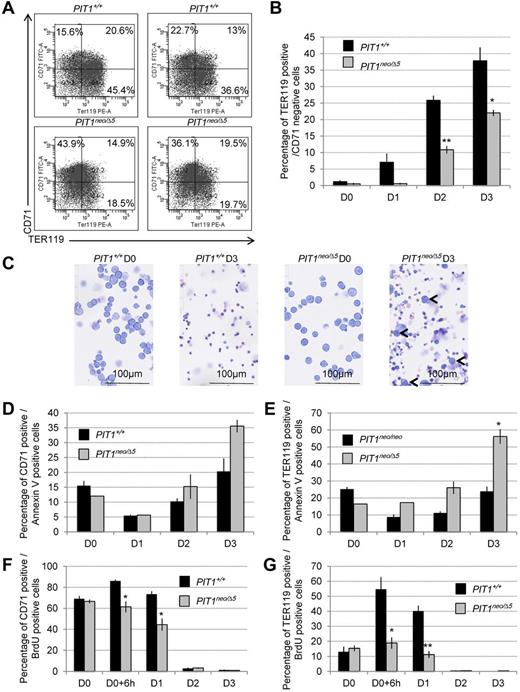
Similar results were obtained during ex vivo maturation of fetal liver cells isolated from PIT1+/+ or PIT1neo/Δ5 littermates. Results showed that when CD71+/TER119+ cells were expressing only 5% of PIT1, few of them could maturate as CD71−/TER119+ cells (Figure 6A-C), suggesting a defect in the terminal maturation of PIT1neo/Δ5 cells. Impaired maturation did not appear to be caused by increased apoptosis (Figure 6D-E), but rather by modification of the cell cycle observed as early as 6 hours after maturation induction, as suggested by BrdU incorporation (Figure 6F-G). These results show that PIT1 expression increases during primary erythroid progenitor maturation and that a low expression of PIT1 impairs erythroid differentiation, leading to the persistence of immature cells.
Fetal liver cells underexpressing PIT1 display erythroid maturation defects. Primary erythroid progenitors were isolated from fetal livers of PIT1+/+ and PIT1neo/Δ5 mice and cultured ex vivo during 4 days. (A) Representative flow cytometric analysis of primary erythroid progenitors 3 days after ex vivo maturation induction. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (B) Percentage of cells that were TER119+/CD71−. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01. (C) May Grünwald-Giemsa stained cytospin of ex vivo cultured erythroid progenitor cells isolated from fetal livers of PIT1+/+ and PIT1neo/Δ5 mice at day 0 (D0) and day 3 (D3). Bar indicates 100μm at a 40× magnification. (D) Percentage of cells that were CD71+/annexin V+, and (E) TER119+/annexin V+ during fetal liver cell ex vivo maturation induction. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05. (F-G) Percentage of fetal liver cells that were CD71+/BrdU+ (F) and TER119+/BrdU+ (G). Cells were cultured with 10μM BrdU for 2 hours and then analyzed 6-14 hours after pulse. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01.
Fetal liver cells underexpressing PIT1 display erythroid maturation defects. Primary erythroid progenitors were isolated from fetal livers of PIT1+/+ and PIT1neo/Δ5 mice and cultured ex vivo during 4 days. (A) Representative flow cytometric analysis of primary erythroid progenitors 3 days after ex vivo maturation induction. Percentages of labeled cells were calculated by taking into account the background labeling (baseline defined by omitting Ab). (B) Percentage of cells that were TER119+/CD71−. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01. (C) May Grünwald-Giemsa stained cytospin of ex vivo cultured erythroid progenitor cells isolated from fetal livers of PIT1+/+ and PIT1neo/Δ5 mice at day 0 (D0) and day 3 (D3). Bar indicates 100μm at a 40× magnification. (D) Percentage of cells that were CD71+/annexin V+, and (E) TER119+/annexin V+ during fetal liver cell ex vivo maturation induction. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05. (F-G) Percentage of fetal liver cells that were CD71+/BrdU+ (F) and TER119+/BrdU+ (G). Cells were cultured with 10μM BrdU for 2 hours and then analyzed 6-14 hours after pulse. Data indicate the means ± SEM of at least 3 independent experiments. Significant differences from PIT1+/+ are indicated. *P < .05; **P < .01.
EKLF and PIT1 expression were concomitantly increased during G1E-ER-Gata1 cell maturation
We analyzed PIT1 and EKLF mRNA (Figure 7A) and protein (Figure 7B) expression in G1E-ER-Gata1 cells treated with β-estradiol for 10, 24 and 33 hours. Results showed an increase in EKLF and PIT1 mRNA and protein expressions during G1E cell maturation. EKLF expression increased before that of PIT1, which is in agreement with our results obtained in primary erythroid progenitor cultures (Figure 5A-B) and consistent with the notion that EKLF could activate PIT1 expression during erythroid maturation.
EKLF-driven PIT1 expression is mandatory for erythroid maturation in vitro. (A) EKLF and PIT1 mRNA quantification during G1E-ER-Gata1 maturation. Means ± SEM of at least 3 experiments are presented. Significant differences from t = 0 are indicated. (B) Western blot of PIT1 and EKLF during G1E-ER-Gata1 maturation. (C) PIT1 and EKLF mRNA levels on murine G1E-ER-Gata1 erythroid cells transduced with lentivirus containing a nontargeting shRNA or shRNAs directed against PIT1 or EKLF. (D) Western blot of PIT1 and EKLF on murine G1E-ER-Gata1–transduced cells. (E-F) PIT1 (E) and EKLF (F) mRNA relative expression in G1E-ER-Gata1–transduced cells 24 hours (EKLF) and 48 hours (PIT1) after maturation induction. Results are normalized to scramble at t = 0 and are expressed as the percentage induction compared with scramble at t = 24 or 48 hours. (G,I) Dematin mRNA (G) and β-globin (I) expression in G1E-ER-Gata1–transduced cells 48 hours after maturation induction. Results are normalized to scramble at t = 0 and are expressed as the percentage induction compared with scramble at t = 48 hours. (H) Percentage of benzidine-positive G1E-ER-Gata1–transduced cells 48 hours after maturation induction. (J) Percentage of benzidine-positive cells 48 hours after maturation induction of EKLF-depleted G1E-ER-Gata1 cells transduced with PIT1 coding lentivirus (PIT1-GFP) or with a control lentivirus (IRES-GFP). (K-L) β-Globin (K) and dematin (L) mRNA expression in G1E-ER-Gata1 cells 48 hours after maturation induction. Results are normalized with control at t = 0 and are expressed as the percentage induction compared with PIT1-GFP at t = 48 hours. Means ± SEM of 5 independent experiments are presented. Significant differences from scramble at t = 48 hours are indicated. *P < .05; **P < .01; ***P < .001.
EKLF-driven PIT1 expression is mandatory for erythroid maturation in vitro. (A) EKLF and PIT1 mRNA quantification during G1E-ER-Gata1 maturation. Means ± SEM of at least 3 experiments are presented. Significant differences from t = 0 are indicated. (B) Western blot of PIT1 and EKLF during G1E-ER-Gata1 maturation. (C) PIT1 and EKLF mRNA levels on murine G1E-ER-Gata1 erythroid cells transduced with lentivirus containing a nontargeting shRNA or shRNAs directed against PIT1 or EKLF. (D) Western blot of PIT1 and EKLF on murine G1E-ER-Gata1–transduced cells. (E-F) PIT1 (E) and EKLF (F) mRNA relative expression in G1E-ER-Gata1–transduced cells 24 hours (EKLF) and 48 hours (PIT1) after maturation induction. Results are normalized to scramble at t = 0 and are expressed as the percentage induction compared with scramble at t = 24 or 48 hours. (G,I) Dematin mRNA (G) and β-globin (I) expression in G1E-ER-Gata1–transduced cells 48 hours after maturation induction. Results are normalized to scramble at t = 0 and are expressed as the percentage induction compared with scramble at t = 48 hours. (H) Percentage of benzidine-positive G1E-ER-Gata1–transduced cells 48 hours after maturation induction. (J) Percentage of benzidine-positive cells 48 hours after maturation induction of EKLF-depleted G1E-ER-Gata1 cells transduced with PIT1 coding lentivirus (PIT1-GFP) or with a control lentivirus (IRES-GFP). (K-L) β-Globin (K) and dematin (L) mRNA expression in G1E-ER-Gata1 cells 48 hours after maturation induction. Results are normalized with control at t = 0 and are expressed as the percentage induction compared with PIT1-GFP at t = 48 hours. Means ± SEM of 5 independent experiments are presented. Significant differences from scramble at t = 48 hours are indicated. *P < .05; **P < .01; ***P < .001.
Knockdown of EKLF decreased PIT1 expression in G1E-ER-Gata1 cells
To confirm whether endogenous EKLF could regulate PIT1 expression and to test the functionality of EKLF interaction with the PIT1 promoter, PIT1 expression was analyzed after transduction of G1E-ER-Gata1 cells with lentivirus expressing shRNA directed against PIT1 or EKLF mRNA. Results showed that PIT1 mRNA and protein expression were decreased when EKLF was knocked down in G1E cells (Figure 7C-D), confirming a regulation of PIT1 expression by EKLF in mouse erythroid cells. Surprisingly, we also found that knockdown of PIT1 decreased basal EKLF expression in G1E-ER-Gata1 cells.
PIT1 reexpression partially restores maturation of G1E-ER-Gata1 cells underexpressing EKLF
To confirm the role of PIT1 during erythroid maturation in an in vitro system, we monitored the maturation of G1E-ER-Gata1 cells transduced with lentivirus-containing shRNAs. Two shRNA targeting independent regions of PIT1 and EKLF mRNAs were used. We first quantified PIT1 (Figure 7E; supplemental Figures 5A, 6A) and EKLF (Figure 7F; supplemental Figures 5B, 6B) gene expression 24 and 48 hours after maturation induction. As expected, PIT1 expression increased to a lesser extent in G1E-containing PIT1 or EKLF shRNAs compared with nontargeting shRNA-transduced cells. The weak increase in PIT1 expression was correlated with the weak increase observed in dematin expression (Figure 7G), a known EKLF target gene,12 arguing for a role of EKLF in regulating PIT1 expression.
Similar to that was observed in primary erythroid progenitors, we used benzidine staining to show that the percentage of hemoglobinized cells after 48 hours of maturation was decreased in cells knocked down for EKLF and in cells underexpressing PIT1 compared with cells transduced with a nontargeting shRNA lentivirus (Figure 7H; supplemental Figure 6C). We confirmed these data by measuring the expression of β-globin 48 hours after maturation induction (Figure 7I; supplemental Figures 5C, 6D). These results strongly suggest that knockdown of PIT1 expression impaired erythroid maturation.
To further confirm a role of PIT1 during erythroid maturation, we monitored the maturation of EKLF-depleted G1E-ER-Gata1 cells transduced with a lentivirus expressing PIT1 along with green fluorescent protein (PIT1-GFP) or with a control lentivirus (GFP). Overexpression of PIT1 in cells knockdown for EKLF partially restored their hemoglobinization, as demonstrated by the percentage of benzidine-positive cells (78.8% in cells overexpressing PIT1 vs 69% in control cells; Figure 7J) and by the increase in β-globin expression (Figure 7K). Moreover, EKLF's direct target dematin was unaffected by PIT1 reexpression (Figure 7L). These results demonstrate that reexpression of PIT1 partially rescues the maturation process of EKLF-depleted erythroid cells and that PIT1 is mandatory for erythroid maturation.
Discussion
In the present study, we addressed the role of PIT1 during hematopoietic development. We have shown that: (1) primary erythroid progenitors down-expressing PIT1 had impaired maturation and (2) the phenotypic features of hematopoietic cells lacking PIT1 were cell autonomous. We also showed that PIT1 was up-regulated during RBC maturation under the control of EKLF, a transcription factor essential for erythroid terminal differentiation. Depleting PIT1 or EKLF from the G1E erythroid cell line led to maturation defects, whereas overexpression of PIT1 in EKLF-depleted G1E cells could partially restore the maturation defects. These results show that a normal expression of PIT1 is critical for RBC terminal differentiation.
Although disruption of PIT1 in mice leads to a dramatic anemia, eventually causing the death of the embryo at midgestation, we previously reported that PIT1-null erythroid progenitors were able to proliferate and differentiate normally in methylcellulose-supported cell culture.6 However, our data illustrated the persistence of embryonic erythrocytes,6 which led us to investigate the role of PIT1 in the maturation of erythrocytes and a possible functional link between PIT1 and EKLF. Indeed, the phenotype of EKLF-KO mice displayed striking similarities to that of PIT1-KO embryos,10,25 such as a midgestation embryonic lethality due to severe anemia, despite the normal proliferation of erythroid progenitors in vitro.12 For both KO mouse models, erythroid cells accumulated at the erythroid progenitor stage and displayed cell membrane defects15 and a globin switch impairment.10,25 Studies of EKLF-KO erythroid cells in vivo and in vitro have shown that EKLF regulates numerous genes involved in hemoglobin metabolism and membrane stability.11-14 We show herein an increased expression of PIT1 during maturation of primary erythroid progenitors and G1E cells that is preceded by an increase of EKLF expression. Our data are in agreement with previous transcriptomic data obtained on G1E-ER4 cells.36 Using a promoter-reporter approach, we documented the regulation of PIT1 by EKLF, but were unable to define precisely the EKLF-binding site in the PIT1 promoter. Although the EKLF consensus binding site 5′CCN CNC CCN3′9 has been redefined more precisely,37 our data argue for the presence of a noncanonical EKLF-binding site in the PIT1 promoter, as was found in the AHSP promoter.38
Despite a molecular link between PIT1 and EKLF and the presence of striking phenotypic similarities between corresponding KO mice, it is noteworthy that PIT1-KO mice die earlier than EKLF-KO mice. This difference could be related to the fetal liver phenotype of E12.5 PIT1-KO embryos, displaying hypoplastic and apoptotic livers.6 Indeed, we cannot exclude that the fetal liver microenvironment could play an additive role in the lethal phenotype of PIT1KO mice. Nevertheless, our present data argue for the existence of a hematopoietic cell-autonomous defect. Adult PIT1neo/neo mice constitutively underexpressing PIT1 and Mx1-Cre;PIT1lox/lox mice specifically depleted from PIT1 in the hematopoietic cell compartment at the adult stage had similar hematopoietic phenotypes. Moreover, ex vivo cultures of primary erythroid progenitors isolated from PIT1neo/neo mice showed impaired maturation compared with PIT1 wild-type cells. BM reconstitution of sublethally irradiated mice experiments showed that, even when placed in a wild-type environment, PIT1KO hematopoietic progenitors could not proliferate and differentiate, demonstrating that the hematopoietic defect in PIT1-null mice arises from a cell-autonomous defect.
In agreement with these data, we showed that depleting PIT1 from G1E-ER-Gata1 cells negatively modulated erythroid maturation. This cell line is an appropriate model to reproduce the physiologic cascade of events that takes place during erythroid maturation.34,35 Even if PIT1 knockdown decreases hemoglobinization of G1E cells and β-globin accumulation, no differences could be observed on TER119 cell-surface marker appearance (not shown) probably because of the incomplete PIT1 down-regulation. Conversely, ex vivo culture of primary erythroid progenitors in which PIT1 was underexpressed showed a clear impaired maturation together with a slight increase in P21 expression. This latter observation is of great interest because a correct balance of progenitor cell proliferation versus lineage-committed differentiation that is defined by the exit from the cell cycle is central to the homeostasis of erythropoiesis. Cell-cycle arrest occurs during the G1 phase as terminal differentiation begins.39,40 It has been found that EKLF can bind directly to P21,16 p18,18 and the E2F2 promoter,15,17 suggesting that the cell-cycle arrest in the G1 phase during erythroid maturation could be because of the direct effects of EKLF. The precise mechanism by which PIT1 affects erythroid maturation is still unknown, although new functions for PIT1 are beginning to emerge. We showed previously that siRNA-mediated depletion of PIT1 expression in HeLa and HepG2 cells reduces cell proliferation and delays the cell cycle.41 Whether these new functions can be involved in the modulation of erythroid maturation requires additional investigations.
In the present study, we show that the contribution of PIT1 to the modulation of erythroid maturation could occur through a modulation of EKLF expression or activity. We documented a decrease in EKLF expression when PIT1 was knocked down, as well as a weak increase of EKLF expression during the maturation of PIT1-depleted G1E-ER-Gata1 cells, suggesting the possible existence of a feedback loop regulating EKLF expression. Overexpression of PIT1 in EKLF-depleted cells partially restores the EKLF expression increase during erythroid maturation, suggesting that PIT1 is instrumental in the EKLF-mediated maturation of erythroid cells. Recent unpublished data in our laboratory have identified the Eleven Nineteen Leukemia (ENL) protein as a protein partner of PIT1. Because ENL is a transcriptional coactivator involved in transcriptional elongation and chromatin modification,42-44 one can hypothesize that PIT1 could regulate EKLF expression indirectly by modulating ENL activity. Additional experiments would be needed to elucidate the precise role of PIT1 in erythroid maturation, but the hypothesis that PIT1 could regulate ENL activity opens new perspectives on PIT1 function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr S. Fabrega (IFR94, Paris) for the generation of lentiviral vector stocks; C. Cordier and J. Megret (IFR94, Paris) for cell sorting; Berissi (IFR94, Paris) for histology; Dr Mitchell Weiss (Children's Hospital, Boston, MA) for the generous gift of G1E-ER-Gata1 cells; Dr M. Pontoglio (Institut Cochin, Paris) and Dr F. Coté (UMR8147, Paris) for helpful discussions; Y. Sassier (Inserm U845, Paris) for technical help; and Dr T. Mercher (Institut Gustave Roussy, Villejuif) and Dr S. Ezine (Inserm U591, Paris) for providing the Mx1-Cre and Rag KO mice, respectively.
This study was supported by grants from Agence Nationale de la Recherche, Inserm, Université Paris Descartes, and Association Laboratoires de Recherches Physiologiques. A.F. and A.R. were supported by the Agence Nationale de la Recherche.
Authorship
Contribution: A.F. designed and performed the research, collected, analyzed, and interpreted the data, performed the statistical analysis, and wrote and reviewed the manuscript; L.B. designed the research and wrote and reviewed the manuscript; C.L., A.R., and V.B. performed the research; I.C. contributed to discussions and reviewed the manuscript; G.C. contributed to discussions; O.H. reviewed the manuscript; and G.F. analyzed and interpreted the data, contributed to discussions, and reviewed the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anne Forand, Centre de Recherche Inserm U845, Université Paris Descartes, Faculté de Médecine Necker, 156 Rue de Vaugirard, 75015 Paris, France; e-mail: anne.forand@inserm.fr.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal