Key Points
CLEC4M plays a role in the clearance of VWF.
CLEC4M polymorphisms contribute to the genetic variability of VWF plasma levels.
Abstract
Genetic variation in or near the C-type lectin domain family 4 member M (CLEC4M) has been associated with plasma levels of von Willebrand factor (VWF) in healthy individuals. CLEC4M is a lectin receptor with a polymorphic extracellular neck region possessing a variable number of tandem repeats (VNTR). A total of 491 participants (318 patients with type 1 von Willebrand disease [VWD] and 173 unaffected family members) were genotyped for the CLEC4M VNTR polymorphism. Family-based association analysis on kindreds with type 1 VWD demonstrated an excess transmission of VNTR 6 to unaffected individuals (P = .0096) and an association of this allele with increased VWF:RCo (P = .029). CLEC4M-Fc bound to VWF. Immunofluorescence and enzyme-linked immunosorbent assay demonstrated that HEK 293 cells transfected with CLEC4M bound and internalized VWF. Cells expressing 4 or 9 copies of the CLEC4M neck region VNTR showed reduced interaction with VWF relative to CLEC4M with 7 VNTR (CLEC4M 4%-60% reduction, P < .001; CLEC4M 9%-45% reduction, P = .006). Mice expressing CLEC4M after hydrodynamic liver transfer have a 46% decrease in plasma levels of VWF (P = .0094). CLEC4M binds to and internalizes VWF, and polymorphisms in the CLEC4M gene contribute to variable plasma levels of VWF.
Introduction
von Willebrand factor (VWF) is a plasma glycoprotein that mediates platelet adhesion and aggregation and acts as the carrier protein for factor VIII (FVIII). VWF synthesis by endothelial cells1 and megakaryocytes2 involves complex post-translational modifications including dimerization, glycosylation, sulfation, multimerization, and propeptide cleavage (reviewed by Sadler3 ). The protein is either constitutively secreted into the plasma and subendothelium or is stored in endothelial Weibel-Palade bodies or platelet α granules from which release can be mediated by a number of chemical and biomechanical stimuli.
Plasma VWF levels in healthy participants show a fourfold range (0.50-2.00 IU/mL).4 These levels are influenced by a variety of genetic and acquired factors. ABO blood group contributes approximately 30% of the genetic influence,5,6 whereas age,6,7 acute-phase stimuli,8-10 and several endocrine abnormalities represent acquired determinants of VWF levels.7,11 In type 1 von Willebrand disease (VWD), which is defined as a partial deficiency of functionally normal VWF, approximately 35% of individuals do not have a putative mutation in the coding region, splice junctions, or proximal promoter of the VWF gene, suggesting that genes other than VWF may contribute to the pathophysiology of this disease.12,13
VWF circulates in a tight, noncovalently linked complex with FVIII. The mean circulating half-lives of VWF and FVIII are 12 to 18 hours and12 hours, respectively, but details of the fate of both proteins are minimal. Evidence exists that cells in the liver and spleen contribute to the clearance of both VWF and FVIII,14 and previous studies have shown that the LDL receptor-related protein (LRP-1) and other members of the LDL receptor family of proteins influence FVIII (reviewed by Lenting et al15 ) and VWF clearance.16 Glycosylation of VWF influences plasma clearance.17 Carbohydrate makes up approximately 20% of the mass of the mature VWF subunit, which includes 12 N-linked high-mannose–containing oligosaccharide chains and 10 O-linked oligosaccharides.18 The N-linked glycans undergo further processing with, among other modifications, the addition of ABO groups, in the post-Golgi compartment.19
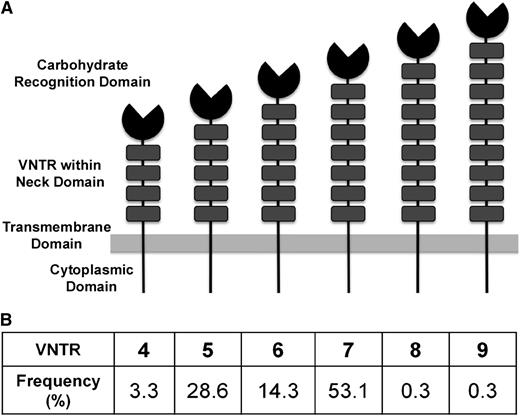
A genome-wide association study (GWAS) meta-analysis identified several novel candidate loci that influence both VWF and FVIII.20 Among these loci, single-nucleotide polymorphisms (SNPs) in or near the C-type lectin receptor, C-type lectin member 4 family M (CLEC4M), showed consistent statistical association with VWF and FVIII levels in both the discovery and replication cohorts. CLEC4M, a calcium-dependent mannose-specific receptor, is expressed by endothelial cells in liver sinusoids and lymph nodes (reviewed by Khoo et al21 ). The receptor has an N-terminal cytoplasmic region, a neck region, and a carbohydrate recognition domain (CRD). The neck region is highly polymorphic, containing 3 to 9 variable numbers of tandem repeats (VNTR) of a conserved 23-amino-acid sequence.22 (Figure 1A) The neck region stabilizes CLEC4M on the endothelial surface by mediating tetramerization of the monomers, and influences the conformation of the CRD.23-25 The CLEC4M VNTR have been implicated in the genetic susceptibility to pathogens such as HIV26 and SARS-CoV.27 CLEC4M has not been previously linked to the clearance of endogenous glycoproteins.
The Structure of CLEC4M and the distribution of CLEC4M VNTR alleles in individuals with type 1 VWD and their families. (A) CLEC4M is organized into 4 domains: an N-terminal cytoplasmic domain, a neck region, and a CRD. The neck region is highly polymorphic, containing VNTR of a highly conserved 23-amino-acid sequence, with a range of 4 to 9 repeats. (B) The distribution of the CLEC4M VNTR alleles as seen in 491 participants with type 1 VWD and their family members is shown.
The Structure of CLEC4M and the distribution of CLEC4M VNTR alleles in individuals with type 1 VWD and their families. (A) CLEC4M is organized into 4 domains: an N-terminal cytoplasmic domain, a neck region, and a CRD. The neck region is highly polymorphic, containing VNTR of a highly conserved 23-amino-acid sequence, with a range of 4 to 9 repeats. (B) The distribution of the CLEC4M VNTR alleles as seen in 491 participants with type 1 VWD and their family members is shown.
Here, we document the ability of CLEC4M to bind and internalize VWF, and to influence in vivo levels of VWF. Additionally, we provide further evidence that polymorphism of the CLEC4M gene contributes to quantitative VWF variation.
Methods
Study population and VWD phenotyping
Index patients with type 1 VWD as well as affected and unaffected family members were included. Index patients with type 1 VWD were defined as individuals with both a personal history of excessive mucocutaneous bleeding and plasma levels of VWF:Ag and/or VWF:RCo between 0.05 and 0.50 U/mL obtained on at least 2 occasions, an RCo:Ag ratio >0.60, and normal multimers. The study was approved by the Research Ethics Board of Queen’s University and for the Zimmerman PPG subjects, by the Institutional Review Boards of each of the participating centers and centrally by the Institutional Review Board of the Medical College of Wisconsin. Informed consent was obtained from all participants in accordance with the Declaration of Helsinki. Laboratory tests for VWF:Ag, VWF:RCo, and FVIII:C were performed at the source clinic attended by the patient according to local methods.13 Genomic DNA was isolated from blood samples collected in EDTA by a salting-out procedure.28 VWF genotyping was performed as described previously.13
Determination of the VNTR genotype and SNP rs868875
The VNTR polymorphism in exon 4 of CLEC4M was genotyped using the same polymerase chain reaction protocol as described previously.29 The genotype of the SNP, rs868875 studied in the CHARGE GWAS, was determined using the polymerase chain reaction-RFLP protocol described previously by Li et al.30 To validate the genotyping results, 10% of the samples were genotyped again by duplicate genotyping experiments.
Statistical methods
Family-based association analysis.
Family-based association testing (FBAT) was performed using FBAT V2.0.331 to test for the association between specific alleles of VNTR and traits. The type 1 VWD phenotype was evaluated as a discrete trait. VWF:Ag and VWF:RCo were evaluated as quantitative traits. An additive genetic model was used. To account for multiple affected individuals in the same family, the empiric variance option was used for all of the traits listed.
Statistical analysis was performed on experiments with an n = 3 or greater using GraphPad InStat3 software (San Diego, CA) for Windows. t tests or one-way analysis of variance with Tukey or Dunnett post hoc analysis were performed using GraphPad InStat software version 3.06 (La Jolla, CA) or SPSS version 16 (IBM, Armonk, NY). Values are expressed as means ± standard error. Figures denote P < .05 with * and P < .001 with **.
Binding of VWF to CLEC4M- and DC-SIGN-Fc chimeras
Binding of VWF to CLEC4M-Fc chimera was measured as previously described with several modifications.32 VWF was coated at 10 µg/mL; Fc-chimera binding was detected with a horseradish peroxidase–conjugated goat anti-human Fc antibody (AbCam, Cambridge, MA). Alternatively, CLEC4M-Fc was coated at 10 µg/mL, and VWF binding was detected with an anti-VWF–horseradish peroxidase antibody (Dako). Human plasma-derived VWF (FVIII-free) was from Haematologic Technologies Inc (Essex Junction, VT), and FVIII depletion was verified by FVIII enzyme-linked immunosorbent assay (ELISA; Affinity Biologicals, Ancaster, Ontario). Recombinant human VWF was produced in 293 cells.33 Fc chimera proteins (10 µg/mL) were preincubated with 1 mg/mL of mannan for 30 minutes at room temperature.
Hepatic expression of CLEC4M
CLEC4M expression was induced through hydrodynamic liver gene transfer as described by Pegon et al.45 The CLEC4M cDNA (7 VNTR) was cloned into the pLIVE vector, and 8- to 12-week-old C57/BL6 mice received 100 µg of plasmid DNA through hydrodynamic injection as described previously by Pruss et al.33 At 4 days after injection, blood was collected by cardiac puncture. Plasma levels of VWF and factor X were measured by ELISA (Dako and Affinity Biologicals, respectively). Hepatic expression of CLEC4M was confirmed with immunohistochemistry studies using a rabbit anti-CLEC4M antibody (Novus Biologicals, Littleton, CO). All animal experiments were approved by the Queen’s University Animal Care Committee.
Results
Study population
A total of 177 families (491 individuals) were included in the study. The population was composed of 177 index patients with type 1 VWD, 141 family members affected with type 1 VWD, and 173 unaffected family members who had no reported bleeding symptoms and/or normal laboratory values. The mean age of the index patients was 22 years (age range, 1-60 years), mean VWF:Ag was 0.41 IU/mL (range, 0.05-0.88 IU/mL), and mean VWF:RCo was 0.35 IU/mL (range, 0.05-0.66 IU/mL). The population was predominately Caucasian (75%). VWF genotyping had been performed on 305 individuals and identified a putative mutation in 151.
Genotyping results
Figure 1B contains a summary of the VNTR allele frequencies. The most frequent alleles were VNTR 5 (28.6%), 6 (14.3%), and 7 (53.1%). The remaining alleles had frequencies of less than 5%. The minor allele frequency (MAF) with a G allele at the SNP rs868875 is 0.300.
Family-based association analysis
FBAT evaluated the association between the discrete trait of type 1 VWD and the quantitative traits VWF:Ag and VWF:RCo vs the CLEC4M VNTR alleles and rs868875. Only the VNTR 5, 6, and 7 are seen frequently; the analysis only examined these 3 alleles (Table 1). There is significant excess transmission of VNTR 6 to unaffected individuals (z score, −2.59; P = .0096); an association of this allele is seen with increased VWF:RCo levels (z score, −2.26; P = .029) with a borderline association seen for increased VWF:Ag levels (z score, −1.72; P = .085). The SNP rs868875 was not significantly associated with the discrete variable, type 1 VWD (z score, 1.52; P = .13), or the quantitative variables of VWF:RCo, (z score, 0.15; P = .88) and VWF:Ag (z score, 0.15; P = .88). The FBAT analysis was repeated, excluding individuals with known putative VWF mutations, and included 137 nuclear families and 332 individuals. In this subgroup, there remains a significant excess transmission of VNTR 6 to unaffected individuals (z score, –2.41; P = .016), an association of this allele with increased VWF:RCo levels (z score, –2.94; P = .0032) and a now significant association for increased VWF:Ag levels (z score, –2.326; P = .020). Finally, when mutation-positive cases are excluded, the minor allele (G) at rs868875 is significantly associated with the discrete trait of type 1 VWD (z score, 2.152; P = .031).
Family-based association test results
| . | . | . | Type 1 VWD . | VWF:RCo . | VWF:Ag . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Markers . | Allele . | Allele frequency . | Family (no.) . | Z score . | P value . | Family (no.) . | Z score . | P value . | Family (no.) . | Z score . | P value . |
| VNTR | 5 | 0.286 | 53 | 1.85 | .064 | 56 | 1.07 | .29 | 56 | 0.84 | .40 |
| 6 | 0.143 | 40 | −2.59 | .0096 | 42 | −2.19 | .029 | 42 | −1.72 | .085 | |
| 7 | 0.531 | 63 | 0.13 | .90 | 65 | 0.32 | .75 | 65 | 0.28 | .78 | |
| rs868875 | G nt | 0.300 | 46 | 1.52 | .13 | 50 | 0.15 | .88 | 50 | 0.15 | .88 |
| . | . | . | Type 1 VWD . | VWF:RCo . | VWF:Ag . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Markers . | Allele . | Allele frequency . | Family (no.) . | Z score . | P value . | Family (no.) . | Z score . | P value . | Family (no.) . | Z score . | P value . |
| VNTR | 5 | 0.286 | 53 | 1.85 | .064 | 56 | 1.07 | .29 | 56 | 0.84 | .40 |
| 6 | 0.143 | 40 | −2.59 | .0096 | 42 | −2.19 | .029 | 42 | −1.72 | .085 | |
| 7 | 0.531 | 63 | 0.13 | .90 | 65 | 0.32 | .75 | 65 | 0.28 | .78 | |
| rs868875 | G nt | 0.300 | 46 | 1.52 | .13 | 50 | 0.15 | .88 | 50 | 0.15 | .88 |
Association analysis of discrete and continuous outcomes (n = 177 families) was performed using an additive genetic model and empiric variance model.
The z score is a standard score. When considering the binary trait, VWD, a positive score indicates excess transmission of that allele to affected individuals, and a negative score indicates a paucity of transmission of that allele to affected individuals. When considering the quantitative traits, VWF:RCo and VWF:Ag, a negative score indicates an association of that allele with higher levels of the trait in question.
Linkage disequilibrium analysis
Supplemental Methods are included. To determine the linkage disequilibrium between the CLEC4M VNTR and the SNP rs868875, we included only unrelated individuals (n = 392) from the type 1 VWD population. Because there are 6 alleles at the VNTR, we generated 6 genotype variables with each indicating the presence of each of the alleles. Only alleles 5, 6, and 7 are frequent, with allele frequencies of 0.306, 0.151, and 0.495, respectively, whereas the SNP (rs868875) has a MAF of 0.315 in this subpopulation. The D′ and r2 values of the SNP with each of the common VNTR alleles 5, 6, 7 are 0.878, 1.00, 0.939 and 0.731, 0.083, 0.422, respectively. The correlation of D′ and r2 depends on the frequency of the alleles in question.34 When the alleles are seen frequently and with similar rates, D′ and r2 are in the same direction, with a larger D′ matching a larger r2. For example, rs868875 MAF is 0.315, and VNTR 5 allele frequency is 0.306, and the D′ is 0.878 and r2 is 0.731. However, in cases where the allele in question is rare and is significantly different from the MAF, D′ and r2 do not correlate: the larger D′, the smaller r2. For example, rs868875 MAF is 0.315, and VNTR 6 is 0.151, and the D′ is 1.00 and r2 is 0.083. Therefore, as per the D′, the SNP is in significant linkage disequilibrium with the VNTR alleles 5, 6, and 7, and the r2 parameter is disregarded because it is dependent on allele frequency differences between the variants.
In vitro binding of VWF to CLEC4M
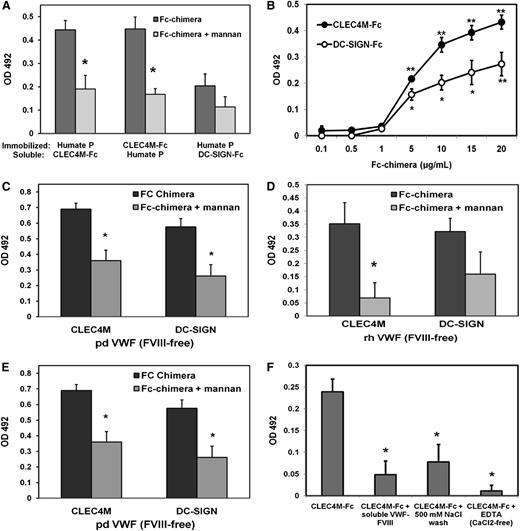
Soluble CLEC4M-Fc bound to immobilized Humate-P (human plasma-derived VWF-FVIII complex) in a dose-dependent manner that saturated at approximately 20 µg/mL CLEC4M-Fc (Figure 2B). Additionally, soluble Humate-P bound to immobilized CLEC4M-Fc (Figure 2A). CLEC4M-Fc also bound to FVIII-free human plasma-derived VWF (Figure 2C) and recombinant human VWF (Figure 2D). Preincubation of CLEC4M-Fc with the mannose polymer, mannan, attenuated binding to all VWF preparations by approximately 50% (Humate P – P = .0072 [Figure 2A]; pdVWF (FVIII-free) – P = .0051 [Figure 2C]; rhVWF – P = .0368 [Figure 2D]). This interaction was calcium dependent, as incubation in calcium-free buffer + EDTA (50 mM) significantly decreased binding, and reversible, as washing with TSM + 500 mM NaCl removed VWF-bound CLEC4M-Fc (Figure 2F). Binding of soluble CLEC4M-Fc to VWF could be competed with soluble Humate-P (20 µg/mL) (Figure 2F). These data demonstrate that CLEC4M binds to VWF alone, as well as to the VWF:FVIII complex. Additionally, DC-SIGN, which is ∼77% homologous to CLEC4M, bound all VWF preparations in a mannose-dependent manner (Figure 2B-D).
Binding of CLEC4M-Fc chimera to VWF. (A-F) Binding of CLEC4M to VWF was measured with a modified ELISA using immobilized VWF or CLEC4M-Fc. Receptors were blocked with mannan (1 mg/mL) for 30 minutes before incubation with VWF. (A) Binding of soluble VWF (10 µg/mL) to immobilized CLEC4M-Fc (10 µg/mL) and soluble CLEC4M-Fc and DC-SIGN-Fc (10 µg/mL) to immobilized Humate-P (10 µg/mL). (B) Dose-response curve of soluble CLEC4M-Fc and DC-SIGN-Fc (0.1-20 µg/mL) binding immobilized Humate-P (1 µg/mL). (C) Binding of CLEC4M and DC-SIGN to FVIII-free plasma-derived VWF, recombinant human VWF (D), and deglycosylated Humate-P (E) was assessed. (F) Calcium dependence, reversibility, and competition for soluble Humate-P were measured. N = 4 to 10 independent experiments; ± SE, *P < .05; **P < .001.
Binding of CLEC4M-Fc chimera to VWF. (A-F) Binding of CLEC4M to VWF was measured with a modified ELISA using immobilized VWF or CLEC4M-Fc. Receptors were blocked with mannan (1 mg/mL) for 30 minutes before incubation with VWF. (A) Binding of soluble VWF (10 µg/mL) to immobilized CLEC4M-Fc (10 µg/mL) and soluble CLEC4M-Fc and DC-SIGN-Fc (10 µg/mL) to immobilized Humate-P (10 µg/mL). (B) Dose-response curve of soluble CLEC4M-Fc and DC-SIGN-Fc (0.1-20 µg/mL) binding immobilized Humate-P (1 µg/mL). (C) Binding of CLEC4M and DC-SIGN to FVIII-free plasma-derived VWF, recombinant human VWF (D), and deglycosylated Humate-P (E) was assessed. (F) Calcium dependence, reversibility, and competition for soluble Humate-P were measured. N = 4 to 10 independent experiments; ± SE, *P < .05; **P < .001.
To examine the contribution of O- and N-glycans to mediating binding of VWF to CLEC4M-Fc, Humate-P was deglycosylated under nondenaturing conditions. Effective de-N- and de-O-glycosylation (which also removed N-glycan terminal sialic acid) of Humate-P was confirmed with concanavalin A and peanut agglutinin-binding assays as described in supplemental Methods (supplemental Data Sets A and B). There was a 70% increase in binding of CLEC4M-Fc to de-O-glycosylated Humate-P (P = .041), and a 75% decrease in binding of CLEC4M-Fc to de-N-glycosylated Humate-P (P = .046) relative to control (Figure 2E). These data demonstrate that CLEC4M binds to VWF in an N-glycan–dependant manner. Binding of CLEC4M and DC-SIGN to rADAMTS13 (supplemental Data Set C) and BSA (data not shown) was used as positive and negative controls, respectively.
Cell-based studies for measuring VWF binding to CLEC4M
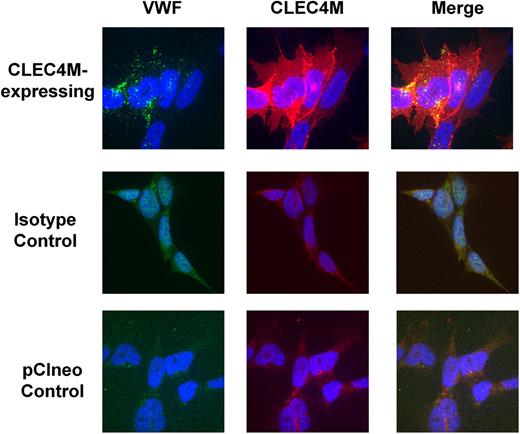
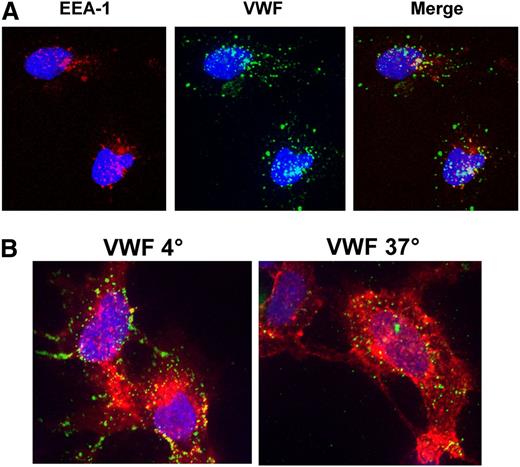
Binding and internalization of VWF by 293 cells stably expressing CLEC4M (7 VNTR allele) was assessed with immunofluorescence and ELISA. VWF binding colocalized with CLEC4M expression, compared with cells transfected with expression vector (pCIneo) and with antibody isotype controls (Figure 3). Preincubation of cells with mannan partially inhibited binding of VWF to CLEC4M-expressing cells (images not shown). VWF and CLEC4M colocalized with early endosomal antigen-1, confirming that CLEC4M mediates internalization of VWF (Figure 4A). Z-stack analysis confirmed that CLEC4M-expressing cells incubated with Humate-P at 4°C retained VWF binding at the cell surface, whereas cells incubated at 37°C had internalized VWF (Figure 4B).
Colocalization of VWF with CLEC4M. To verify that VWF binding colocalized with CLEC4M expression on the cell surface, CLEC4M-expressing cells were incubated with Humate-P (2 U/mL VWF) for 30 minutes, fixed, and labeled for CLEC4M (red), VWF (green), and DAPI (blue). Colocalization of VWF with CLEC4M is observed in yellow (merge). Images are representative of 3 to 4 separate experiments.
Colocalization of VWF with CLEC4M. To verify that VWF binding colocalized with CLEC4M expression on the cell surface, CLEC4M-expressing cells were incubated with Humate-P (2 U/mL VWF) for 30 minutes, fixed, and labeled for CLEC4M (red), VWF (green), and DAPI (blue). Colocalization of VWF with CLEC4M is observed in yellow (merge). Images are representative of 3 to 4 separate experiments.
Internalization of VWF by CLEC4M-expressing cells. (A) Internalization of VWF by CLEC4M-expressing cells was confirmed by demonstrating colocalization of VWF (green) and early endosomal antigen 1 (red). Blue represents the DAPI nuclear counterstain. (B) Incubation of CLEC4M-expressing cells (red) at 4°C maintained VWF (green) at the cell surface, whereas incubation at 37°C facilitated internalization of VWF.
Internalization of VWF by CLEC4M-expressing cells. (A) Internalization of VWF by CLEC4M-expressing cells was confirmed by demonstrating colocalization of VWF (green) and early endosomal antigen 1 (red). Blue represents the DAPI nuclear counterstain. (B) Incubation of CLEC4M-expressing cells (red) at 4°C maintained VWF (green) at the cell surface, whereas incubation at 37°C facilitated internalization of VWF.
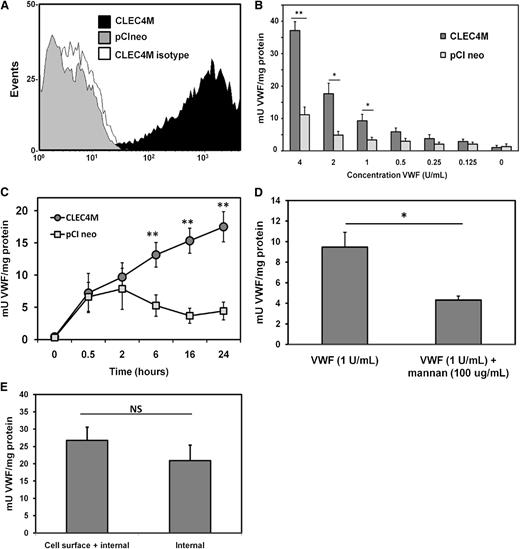
Flow cytometry was used to confirm CLEC4M expression by stable cell lines (Figure 5A). CLEC4M-expressing cells bound and internalized VWF in a dose-dependent (Figure 5B) and time-dependent (Figure 5C) manner relative to controls stably transfected with expression vector pCIneo. Preincubation of CLEC4M-expressing cells with mannan inhibited VWF binding and internalization by approximately 50% (P = .0088, Figure 5D). Internalization of VWF by CLEC4M-expressing cells was confirmed by removing cell surface VWF with trypsin at the end of the VWF incubation period to cleave the CRD (VWF binding) of CLEC4M. Levels of internalized VWF were compared between non–trypsin-treated cell lysates with cell surface internalized VWF; no significant differences were observed (P = .3587, Figure 5E). These studies confirm that CLEC4M binds to VWF on the cell surface, and that the VWF-CLEC4M complex undergoes receptor-mediated endocytosis.
Quantification of VWF internalization by CLEC4M-expressing cells. (A-E) To quantify binding and internalization of VWF by cells stably expressing CLEC4M, cells were incubated with Humate-P, washed, and lysed. Levels of VWF in the cell lysate were quantified by ELISA. (A) Flow cytometric confirmation of CLEC4M expression by stable cell lines. (B-C) Levels of VWF in the cell lysate were measured as a dose response and as a time course. (D) Preincubation of CLEC4M-expressing cells with mannan (100 µg/mL) partially attenuated VWF binding and internalization. (E) Internalization of VWF by CLEC4M was confirmed by incubating CLEC4M-expressing cells with 2 U of VWF for 24 hours, and measuring levels of VWF in cell lysates prepared from trypsinized and nontrypsinized cells. N = 4 to 6 independent experiments; ± SE, *P < .05; **P < .001.
Quantification of VWF internalization by CLEC4M-expressing cells. (A-E) To quantify binding and internalization of VWF by cells stably expressing CLEC4M, cells were incubated with Humate-P, washed, and lysed. Levels of VWF in the cell lysate were quantified by ELISA. (A) Flow cytometric confirmation of CLEC4M expression by stable cell lines. (B-C) Levels of VWF in the cell lysate were measured as a dose response and as a time course. (D) Preincubation of CLEC4M-expressing cells with mannan (100 µg/mL) partially attenuated VWF binding and internalization. (E) Internalization of VWF by CLEC4M was confirmed by incubating CLEC4M-expressing cells with 2 U of VWF for 24 hours, and measuring levels of VWF in cell lysates prepared from trypsinized and nontrypsinized cells. N = 4 to 6 independent experiments; ± SE, *P < .05; **P < .001.
Contribution of CLEC4M genetic variability to VWF binding and internalization
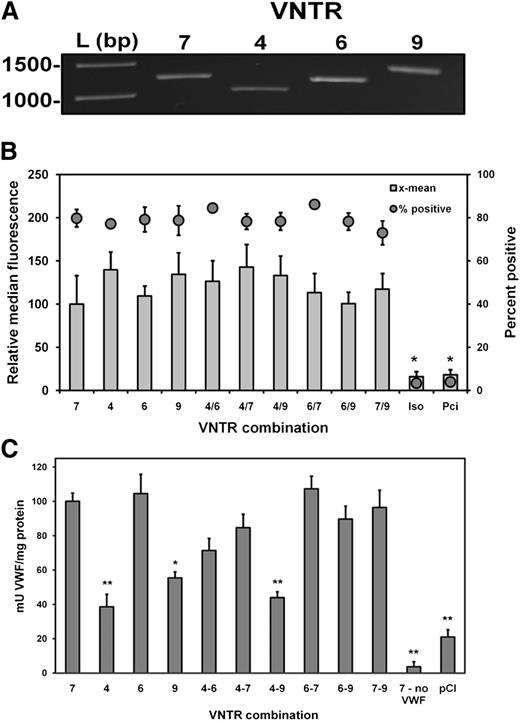
The contribution of the CLEC4M VNTR to differential VWF binding and internalization was measured on 293 cells expressing CLEC4M with 4, 6, 7, and 9 tandem repeats (Figure 6A). Expression levels of CLEC4M variants by transfected 293 cells was quantified by flow cytometry (Figure 6B). Decreased binding and internalization of VWF was observed with cells expressing CLEC4M 4 and 9 tandem repeats compared with CLEC4M with 7 tandem repeats (CLEC4M 4 allele – 60% reduction, P < .001; CLEC4M 9 allele – 45% reduction, P = .006, Figure 6C). Cells expressing the VNTR construct combination 4 and 9 had a 55% decrease in binding and internalization of VWF relative to cells expressing CLEC4M with 7 VNTR (P < .001), whereas cells expressing CLEC4M combinations 4/6 and 4/7 had reduced, but not statistically significant, VWF binding and internalization (CLEC4M 4/6 – P = .157, CLEC4M 4/7 – P = .787). No difference in binding and internalization of VWF was seen in cells expressing 6 tandem repeats compared with cells with the 7 allele. Differences in median fluorescence did not account for variances in VWF binding and internalization. Normalization of VWF in cell lysates to the levels of CLEC4M expression did not significantly influence the observed pattern of VWF binding and internalization (supplemental Data Set D).
Contribution of CLEC4M tandem neck region polymorphisms to VWF binding and internalization. (A) CLEC4M variable tandem repeat constructs (4, 6, and 9) were created. (B) A total of 293 cells were transiently transfected with constructs singly or in combinations, and CLEC4M expression was verified with flow cytometry. (C) Duplicate transfection conditions were exposed to VWF (1 U/mL) for 24 hours. Cells were washed and lysed, and levels of VWF in the cell lysate were quantified by ELISA. N = 6 to 8 independent experiments; ± SE, *P < .05; **P < .001.
Contribution of CLEC4M tandem neck region polymorphisms to VWF binding and internalization. (A) CLEC4M variable tandem repeat constructs (4, 6, and 9) were created. (B) A total of 293 cells were transiently transfected with constructs singly or in combinations, and CLEC4M expression was verified with flow cytometry. (C) Duplicate transfection conditions were exposed to VWF (1 U/mL) for 24 hours. Cells were washed and lysed, and levels of VWF in the cell lysate were quantified by ELISA. N = 6 to 8 independent experiments; ± SE, *P < .05; **P < .001.
Murine hepatic expression of CLEC4M
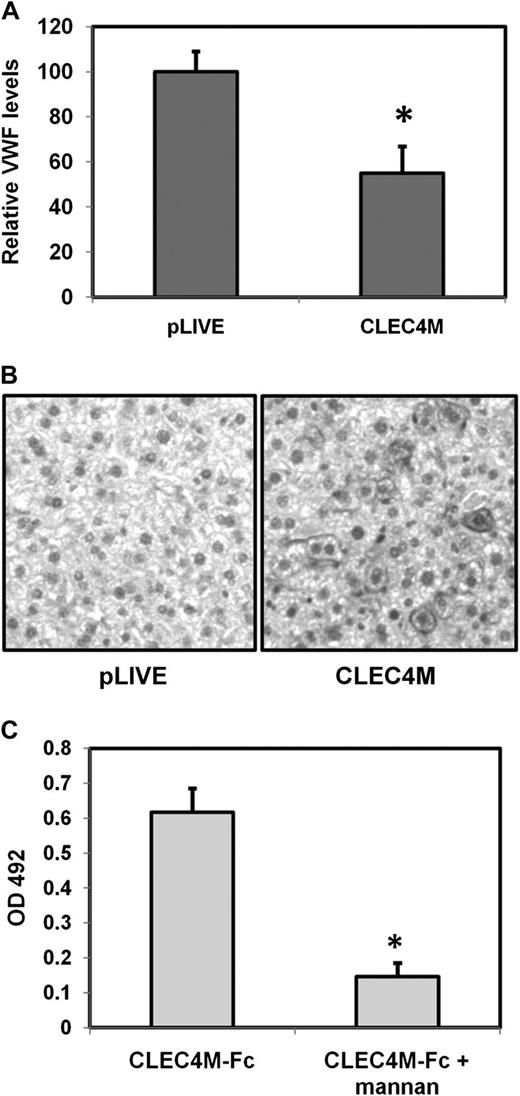
As mice do not express a CLEC4M ortholog, hepatocytic expression of CLEC4M was induced using hydrodynamic gene transfer, with the empty pLIVE vector administered to control mice. At 4 days after injection, blood and liver tissues were collected. Hepatic CLEC4M expression was assessed through immunohistochemistry (Figure 7B). Mice in which 10% to 30% of hepatocytes are expressing CLEC4M showed a 46% decrease in plasma levels of VWF (P = .0094) (Figure 7A). However, no significant difference in plasma levels of the liver-synthesized glycoprotein factor X were observed (P = .33), suggesting that hydrodynamic gene transfer does not significantly disrupt hepatic protein synthesis. Additionally, CLEC4M-Fc bound murine recombinant VWF in a glycan-dependent manner (Figure 7C), suggesting that liver expression of CLEC4M accelerates VWF clearance.
Hepatic expression of CLEC4M is associated with decreased plasma levels of VWF. (A) Plasma levels of VWF in mice expressing CLEC4M in >10% hepatocytes (N = 16). (B) Confirmation of CLEC4M expression by immunohistochemistry studies. (C) CLEC4M-Fc binds recombinant murine VWF in a glycan-dependent manner. ± SE, *P < .05.
Hepatic expression of CLEC4M is associated with decreased plasma levels of VWF. (A) Plasma levels of VWF in mice expressing CLEC4M in >10% hepatocytes (N = 16). (B) Confirmation of CLEC4M expression by immunohistochemistry studies. (C) CLEC4M-Fc binds recombinant murine VWF in a glycan-dependent manner. ± SE, *P < .05.
Discussion
Although the biosynthetic control of VWF has been explored in many studies, very little is known about where and how it is cleared from the circulation. In the CHARGE GWAS meta-analysis, 6 novel loci were identified to be associated with the plasma levels of VWF in healthy participants.20 The function of one of these loci, CLEC4M, a calcium-dependent, mannose-specific receptor, suggests a potential role in VWF clearance.
FBAT analysis compares the observed distribution of traits, either binary (VWD or unaffected) or continuous variables (VWF:Ag and VWF:RCo) and genotypes (both single alleles and more complex genotypes) to expected distributions that are calculated using family data in an additive genetic model. The SNP rs868875 had been the most statistically significant SNP (P = 1.3 × 10−10) identified within the CLEC4M gene in the CHARGE GWAS meta-analysis with the minor allele (G) being associated with lower VWF levels.20 In our population, using FBAT analysis, no association was seen between rs868875 and the binary trait of VWD or the quantitative traits of VWF:RCo and VWF:Ag (Table 1). When mutation-positive cases are excluded, the minor allele (G) at rs868875 was significantly associated with the discrete trait of type 1 VWD (z score, 2.152; P = .031) but remained nonsignificant for VWF:RCo and VWF:Ag. Thus, the effect of the rs868875 SNP in the mutation-negative subgroup is consistent with those reported in the CHARGE GWAS.
The polymorphic CLEC4M neck region (Figure 1A) mediates tetramerization of CLEC4M monomers and influences the conformation of the CRD.24,25 Alleles 5, 6, and 7 account for 96.0% of the alleles seen in our population (Figure 1B). The VNTR 5, 6, and 7 alleles are in linkage disequilibrium with rs868875, suggesting that the association of rs868875 with the VWF:Ag levels may be a reflection of the linkage disequilibrium to the CLEC4M VNTR. Using FBAT analysis (Table 1), the VNTR 6 is associated with the trait of VWD (z score, −2.59; P = .0096) and with the continuous variable of VWF:RCo (score, −2.12; P = .029). A trend was seen with VWF:Ag (z score, −1.72; P = .085). The negative z score in this analysis indicates a paucity of transmission of the VNTR 6 allele to individuals with VWD. When considering the quantitative traits, VWF:RCo and VWF:Ag, the negative score indicates an association of VNTR6 with higher levels of VWF:RCo and VWF:Ag. When the FBAT was repeated excluding the confounding variable of individuals with known putative VWF mutations, there remained a significant excess transmission of VNTR 6 to unaffected individuals (z score, −2.41; P = .016), an association of this allele with increased VWF:RCo levels (z score, −2.94; P = .0032), and now a significant association seen with increased VWF:Ag levels (z score, −2.326; P = .020). Thus, VNTR 6 is associated with reduced risk for VWD and higher VWF activity and VWF levels. These studies provide further evidence that polymorphisms in the CLEC4M gene contribute to quantitative VWF traits. The rare VNTR alleles 4, 8, and 9 were seen too infrequently in our population to be included in the association analysis.
The in vitro and in vivo models presented in our study suggest that CLEC4M functions as a VWF clearance receptor by binding VWF and facilitating its internalization and targeting to early endosomes. As there is no murine CLEC4M ortholog, hepatic expression of CLEC4M was induced in healthy mice through hydrodynamic gene transfer. Although CLEC4M expression is localized primarily to the sinusoidal endothelial cells of the liver and spleen, hepatocyte-mediated VWF clearance has been identified in the context of infection37 and thus may serve as an appropriate surrogate. CLEC4M expression (hepatocytes 10%-30% positive) was associated with decreased plasma levels of VWF:Ag (P = .0094), although plasma levels of the liver-synthesized glycoprotein factor X remained unaltered (P = .33). This finding suggests that CLEC4M can bind to and selectively clear VWF from a complex plasma environment. However, this does not exclude the possibility that additional VWF receptors coexpressed on hepatocytes may enhance the CLEC4M-mediated uptake of VWF by these cells.
In a modified ELISA, recombinant CLEC4M-Fc chimera bound to plasma-derived VWF-FVIII complex (Humate-P) in a dose-dependent manner (Figure 2B) and was variably blocked by competitive inhibition with mannan (Figure 2A,C,D).35 Binding specificity was confirmed using FVIII-free human plasma-derived VWF (Figure 2C) and recombinant human VWF (Figure 2D). The mature VWF subunit is heavily glycosylated with 12 N-linked high-mannose-containing oligosaccharide chains and 10 O-linked oligosaccharides.18 These glycans are required for proper synthesis and secretion of VWF,36 regulation of proteolysis by ADAMST13,37,38 and interaction of VWF with its various ligands.39 VWF glycosylation also influences clearance with the addition of ABO blood groups to N-linked glycan chains being a major determinant of plasma VWF levels.5,19 The addition of A or B glycans is associated with longer VWF survival.17,40 Our data confirm that CLEC4M binds to VWF in a N-linked glycan-dependant manner, with a 75% decrease in binding of CLEC4M-Fc to de-N-glycosylated Humate-P (P = .046) relative to control (Figure 2E). The role of O-linked glycans in VWF clearance is less clear.41,42 De-O-glycosylated VWF demonstrated increased binding to CLEC4M, which is consistent with previously published in vivo data and indicates that O-linked glycosylation has a protective effect against clearance.41
CLEC4M and its homolog, DC-SIGN (dendritic cell-specific intracellular adhesion molecules [ICAM-3] grabbing nonintegrin) are closely related C-type lectins encoded by genes on chromosome 19p13. The 2 proteins show 77% amino acid identity and are similar in structure with a cytoplasmic tail, a coiled neck region made up of a 23-amino-acid repeat region and a mannose-specific carbohydrate domain.43 Both proteins have similar ligand profiles that include ICAM-3 and viral glyoproteins such as HIV-1 gp-120,22 HCV E2,44 and SAR-CoV.27 Our results demonstrating that DC-SIGN also binds VWF and is inhibited by mannan are not surprising (Figure 2B-D). The degree to which either of these proteins contributes to VWF clearance remains to be defined. There are likely a number of additional receptors (at least some of which will bind to glycan-associated structures) that are also involved in VWF clearance. This fact is emphasized by the recent demonstration that Siglec-5, a macrophage-expressed lectin receptor, also binds to and internalizes VWF and FVIII.45 In contrast to CLEC4M, which is expressed on endothelial cells within the liver sinusoids and lymph nodes,22 DC-SIGN is expressed on dendritic cells.46 The VNTR within the neck region of DC-SIGN is much less polymorphic than in CLEC4M, with only 6 to 8 repeats having been identified and the frequency of the VNTR allele 7 being greater that 99%.29 Therefore, although we have demonstrated that DC-SIGN binds VWF and may play a role in VWF clearance, we currently have no evidence that this receptor contributes to the genetic variability of VWF levels. Analysis of DC-SIGN and CLEC4M demonstrates that the 2 genes are not in strong linkage disequilibrium (supplemental Data Set E) and suggests that the associated SNP identified in the CHARGE GWAS study was detecting CLEC4M and not DC-SIGN.
CLEC4M contributes to viral infectivity either in cis by receptor-mediated endocytosis47 or trans by presenting the virus to circulating T cells or peripheral blood mononuclear cells,48 depending on the ligand (reviewed by Lozach et al49 ). In our cell-based studies, CLEC4M binds VWF in a dose-dependent and time-dependent manner (Figure 5B-C). The colocalization of VWF and CLEC4M with early endosomes (Figure 4A) suggests that VWF is endocytosed by CLEC4M-expressing cells. This finding is supported by comparing cell lysates that had been treated with trypsin at the end of the VWF incubation period, to cleave the CRD (VWF binding) of CLEC4M, to lysates from cells not exposed to trypsin (Figure 5E). No significant differences were observed (P = .36) in these studies. Thus, CLEC4M binds to VWF on the cell surface, and the VWF-CLEC4M complex undergoes receptor-mediated endocytosis.
The CLEC4M neck forms extended stalks that are stabilized by lateral interactions of α-helical regions in the area of the VNTR.25 The VNTR region is important in tetramerization of CLEC4M on the cell surface. CLEC4M tetramers are necessary for high-avidity binding to ligands.25 Also, the neck domain and the composition of the VNTR included in the tetramer will determine the conformation of the CRD.24 The effect of the number of VNTR in the neck region on binding of ligands through the CRD and the composition of tetramers made up of homozygous VNTR alleles vs heterozygous alleles is controversial. It is not clear whether certain VNTR alleles or hetero-oligomer formation changes the affinity of CLEC4M binding to ligands.24,50 Conflicting studies report that a minimal number of VNTR is necessary for tetramerization,25 whereas others have reported that VNTR alleles 5, 6, and 7 tetramerize with comparable efficiency.50 HEK293 cells were transfected with VNTR 7 (the most common), 6 (the significant VNTR in the association studies), and 4 and 9 (the lowest and highest VNTR alleles seen in our population). Nonsignificant differences in the expression levels of the CLEC4M VNTR variants (Figure 6B) did not account for variances in VWF binding and internalization. Cells expressing the VNTR 4 and 9 had a 55% decrease in binding and internalization of VWF relative to cells expressing CLEC4M with 7 VNTR (P < .001, Figure 6C). Unfortunately, the allele frequency for VNTR 4 was 3.3% and for VNTR 9 was 0.3% in our population, making any meaningful association analysis difficult. On the other hand, the cells expressing VNTR 6 demonstrated comparable VWF binding and internalization to cells transfected with VNTR 7. This finding is in conflict with our human data that found an association of VNTR 6 with a normal phenotype, and increased VWF:RCo and VWF:Ag levels. Thus, the exact mechanism by which the CLEC4M VNTR may affect the function of this receptor remains unclear and is consistent with previous reports where similar conflicting findings have been reported.24,25,50
A major limitation of this model is that subtle and relevant in vivo factors are not accounted for. One of these influences is shear stress mediated by the flowing blood that will affect VWF conformation. The finding that CLEC4M’s CRD is hinged, or flexibly linked to the neck region, further suggests that shear stress may have a significant impact on the interaction of VWF and CLEC4M.25
This investigation has extended previous genetic association evidence implicating the CLEC4M locus as a determinant of VWF plasma levels, and has shown, through a number of complementary animal model, cell, and molecular biology approaches that CLEC4M binds VWF and subsequently mediates its internalization. We have also demonstrated that the CLEC4M VNTR polymorphism mediates differences in VWF binding and internalization in vitro. Whether this influence also contributes to the variability of plasma VWF levels is unclear and warrants further investigation.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the clinical contribution of Dr Victor Blanchette, from The Hospital for Sick Children, Toronto, Canada, and Dr. Pingzhao Hu for statistical assistance, as well as Matthew Gordon, Jeff Mewborn, and Lee Boudreau from Queen’s University for technical assistance.
This research was supported by grants from CIHR (Canadian Institute of Health Research: grant MOP 97849), the Heart and Stroke Foundation of Canada (grant NA 6881), the CTAQ (Clinical Teachers’ Association of Queen’s University) Endowment Fund, and the Zimmerman Program for the Molecular and Clinical Biology of VWD (HL081588). L.L.S. is the recipient of a Heart and Stroke Foundation of Canada Research Fellowship, a Canadian Hemophilia Society fellowship, and a Queen’s University Senate Advisory Research Council award. N.R. is the recipient of a Bayer Hemophilia Awards Program Clinical Training Award. D.L. is the recipient of a Canada Research Chair in Molecular Hemostasis.
Authorship
Contribution: N.R., L.L.S., C.N., J.J.R. K.S., and B.B. performed the experiments; R.R.M. contributed patient data and DNA samples; A.D.P. performed the FBAT analysis; N.R., L.L.S., and D.L. wrote the paper; and P.D.J. and D.L. designed the research study and edited the paper.
Conflict-of-interest disclosure: P.D.J. receives research funding from CSL Behring and honoraria from CSL Behring, Bayer, and Baxter for educational presentations. The remaining authors declare no competing financial interests.
Correspondence: David Lillicrap, 88 Stuart Street, Richardson Laboratory, Queen’s University, Kingston, Ontario, K7L 3N6, Canada; e-mail: lillicrap@cliff.path.queensu.ca.
References
Author notes
N.R. and L.L.S. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal