Key Points
PEAR1 is a negative regulator of megakaryocyte proliferation in vitro and thrombocyte formation in vivo.
PEAR1 regulates the PI3K/PTEN pathway.
Abstract
Platelet endothelial aggregation receptor-1 (PEAR1) participates in platelet aggregation via sustaining αIIbβ3 activation. To investigate the role of PEAR1 in platelet formation, we monitored and manipulated PEAR1 expression in vitro in differentiating human CD34+ hematopoietic stem cells and in vivo in zebrafish embryos. PEAR1 expression rose during CD34+ cell differentiation up to megakaryocyte (MK) maturation. Two different lentiviral short hairpin knockdowns of PEAR1 did not affect erythropoiesis in CD34+ cells, but increased colony-forming unit MK cell numbers twofold vs control in clonogenic assays, without substantially modifying MK maturation. The PEAR1 knockdown resulted in a twofold reduction of the phosphatase and TENsin homolog (PTEN) phosphatase expression and modulated gene expression of several phosphatidylinositol 3-kinase (PI3K)-Akt and Notch pathway genes. In zebrafish, Pear1 expression increased progressively during the first 3 days of embryo development. Both ATG and splice-blocking PEAR1 morpholinos enhanced thrombopoiesis, without affecting erythropoiesis. Western blots of 3-day-old Pear1 knockdown zebrafish revealed elevated Akt phosphorylation, coupled to transcriptional downregulation of the PTEN isoform Ptena. Neutralization by morpholinos of Ptena, but not of Ptenb, phenocopied the Pear1 zebrafish knockdown and triggered enhanced Akt phosphorylation and thrombocyte formation. In summary, this is the first demonstration that PEAR1 influences the PI3K/PTEN pathway, a critical determinant of Akt phosphorylation, itself controlling megakaryopoiesis and thrombopoiesis.
Introduction
Platelet endothelial aggregation receptor-1 (PEAR1) is a type-1 membrane protein with an EMI domain (characteristic for EMILIN protein family), epidermal growth factor-like repeats, and multiple cytoplasmic tyrosines and prolines.1,2 We recently reported that interactions between PEAR1, Src family kinases (C-src and Fyn), and the p85/phosphatidylinositol 3-kinase 3-kinase (PI3K) subunit constitute a signaling complex, causing sustained αIIbβ3 activation via Akt phosphorylation.1 In addition to its role in platelet function, a potential role for PEAR1 in hematopoietic cell regulation has been proposed.3 The stable overexpression of PEAR1 in mouse bone marrow cells using retroviral vectors decreased the number of myeloid progenitors during in vitro clonogenic assays, suggesting that PEAR1 regulates the early stages of hematopoietic differentiation.3 Whether PEAR1 is also a determinant of megakaryocyte (MK) differentiation is presently unclear.
Thrombopoiesis generates approximately 1011 platelets daily via cytoplasmic fragmentation of MKs matured from bone marrow precursor cells. Megakaryopoiesis and MK differentiation are complex processes controlled by a series of transcription factors and regulated by a variety of cytokines and chemokines.4,5 Differentiation requires a dynamic endomitotic spindle formation, the maturation of a demarcation membrane system, mechanically controlled organelle transport, and microtubule assembly in MKs.6,7
PI3K and phosphatase and TENsin homolog (PTEN) are both tightly linked to Akt signaling, critically controlling proliferation, survival, and migration in hematopoietic cells.8,9 PTEN is a dual-specificity protein phosphatase and an inositol phospholipid phosphatase that dephosphorylates phosphatidylinositol 3,4,5-trisphosphate (PIP3), thus producing phosphatidylinositol 4,5-bisphosphate (PIP2) and thereby negatively regulating oncogenic and nononcogenic PI3K/Akt signaling.10 Indeed, the constitutive activation of PI3K/Akt signaling promotes the uncontrolled growth of hematopoietic cells in commonly detected myeloproliferative disorders and lymphomas.9 PTEN inactivation appears to be one of the major causes of the constitutive activation of PI3K/Akt. Reduced PTEN expression and/or activity in myeloid and lymphocytic leukemia have been linked to mutations, epigenetic repression, or posttranslational inhibition.10 Pten deletion in mouse hematopoietic stem cells (HSCs) predisposes to myeloproliferative disorders. Furthermore, increased megakaryopoiesis is a characteristic of Pten-deficient mice, which have 25% more platelets.11
In view of the tight link between PEAR1 and PI3K activation,1 we have investigated the role of PEAR1-triggered Akt phosphorylation in MK differentiation and thrombopoiesis. Using PEAR1 knockdown strategies in vitro in cell cultures and in vivo in zebrafish, we demonstrate a role for PEAR1 in MK precursor proliferation and thrombocyte formation via regulation of the PI3K/PTEN pathway.
Materials and methods
Human CD34+ differentiation in vitro
Human CD34+ HSCs were separated by magnetic cell sorting from buffy coats and isolated from healthy donor peripheral blood (Miltenyi Biotec, Auburn, CA). Isolated CD34+ cells were cultured in StemSpan SFEM medium (Stem Cell Technologies, Vancouver, BC, Canada), supplemented with 20 ng/mL thrombopoietin (TPO), 10 ng/mL stem cell factor, 10 ng/mL interleukin (IL)-6, and 10 ng/mL FMS (proto-oncogene)-like tyrosine kinase 3 (FLT-3), all from Peprotech (London, United Kingdom). On day 6 of the culture, the following cytokines were resupplemented: 10 ng/mL TPO, 10 ng/mL IL-6, and 10 ng/mL IL-1β. The use of human bone marrow sections for research purposes was authorized by the Ethical Committee of the Academic Hospitals of the KU Leuven, and this study was conducted in accordance with the Declaration of Helsinki. All animal protocols were approved by the Ethical Committee of the KU Leuven.
Inhibition of PEAR1 expression by siRNA
Two short interfering RNA (siRNA) sequences for human PEAR1, GCACGCTGCTCATGTGAAA (referred to subsequently as shPEAR1-1461) and GCAGCTACATGGAGATGAA (shPEAR1-2938), developed using siSearch software (http://www.dharmacon.com/DesignCenter/DesignCenterPage.aspx) were validated via quantitative polymerase chain reaction (PCR) measurements (see the following sections) for their potency to suppress messenger RNA (mRNA) expression of PEAR1 on endothelial EA.hy926 cells. Their number refers to the respective positions in the reference human PEAR1 mRNA (NM_001080471.1) that are recognized by the siRNA.
Generation of lentiviral transfer plasmids
Lentiviral vectors encoding microRNA 30 (miR30)-based knockdown hairpins derived from the aforementioned siRNAs were generated for stable knockdown as previously described12 (referred to as miR_PEAR1_1461 and miR_PEAR1_2938) together with control hairpins directed against the monomeric red fluorescent protein from Discosoma corallimorpharian, DsRed (miR_DsRed). All constructs (pGAE-SFFV (spleen focus forming virus)-BsdR-miR_PEAR1_1461-WPRE (woodchuck hepatitis virus posttranscriptional regulatory element); pGAE-SFFV-BsdR-miR_PEAR1_2938-WPRE, and pGAE-SFFV-BsdR-miR_DsRed-WPRE) contain a blasticidine resistance cassette (BsdR) driven from a SFFV long terminal repeat promoter, followed by the respective miRs. The optimized simian immunodeficiency virus (SIV) vector pGAE was a kind gift from Dr D. Nègre (Ecole Normale Superieure Lyon, France). All cloning steps were sequence verified.
Lentiviral vector production and transduction
Lentiviral vector production was performed essentially as described earlier.13 Briefly, vesicular stomatitis virus glycoprotein pseudotyped lentiviral vector particles were produced by triple transient polyethylenimine transfection in HEK293T cells using pMDG.2, which encodes the vesicular stomatitis virus glycoprotein envelope, pAD_SIV3+, packaging plasmid, and transfer plasmids pGAE-SFFV-BsdR-miR_PEAR1_1461-WPRE and pGAE-SFFV-BsdR-miR_PEAR1_2938 -WPRE, to generate LV_miR_PEAR1_1461 and LV_miR_PEAR1_2938, respectively. Likewise controls with miR-based hairpins that target the mRNA of DsRed were generated, resulting in LV_miR_DsRed. The latter vector will be referred to as “control” throughout the text.
Stable PEAR1 knockdown cells and controls were generated following lentiviral transduction. Typically, CD34+ cells (5 × 105) were transduced twice in 96-well plates (on the day of isolation and the day after).
Cell proliferation and CFU assays
Colony-forming unit (CFU)-MK and CFU-E (erythrocyte colonies) potential was assessed as previously described. Human CD34+ cells (5 × 103) were cultured in Megacult-C 04973 and supplemented with recombinant cytokines according to the manufacturer's instructions. The number of CFU-MK and CFU-E were determined in a semisolid culture system using a commercially available kit (StemCell Technologies). The total number of colonies was counted at 6 and 10 days using a light microscope (Leica DM RBE; Wetzlar, Germany) in cultures. Experiments were carried out in triplicate.
PCR and real-time qRT-PCR
Total mRNAs were extracted with TRIzol (Invitrogen, Carlsbad, CA) from zebrafish embryos or with the Qiagen kit (RNeasy mini kit) from cultured cells. Complementary DNAs were synthesized using M-MLV reverse transcriptase (Invitrogen).
Human gene expression was measured using FAM-labeled TaqMan assay products (Applied Biosystems, Life Technologies, Gent, Belgium) or via regular SYBR Green RT-PCR (IDT, Leuven, Belgium). Zebrafish gene expression was measured using an SYBR Green assay developed for zebrafish. All genes were normalized to the housekeeping genes human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or β-actin (ACTB), zebrafish β-actin (Actb), or elfa. Reverse transcription (RT)-PCR products and primer sequences are listed in the data supplement (supplemental Tables 1 and 2). Quantitative RT (qRT) PCR reactions were analyzed using the ABI 7000 real-time PCR machine (Life Technologies). Expression was quantified via the ΔΔCt method14 and expressed in arbitrary units (defined in figure legends) or in percentage compared with control. Expression of genes included in the PI3K/Akt (reference PAHS-058A) and Notch (reference PAHS-059A) RT2 Profiler signaling was analyzed by qRT-PCR (SABioscience, Qiagen). Cell populations transduced by control lentivirus were used as a reference for LV_miR_PEAR1-transduced cells.
Western blot analysis
Western blot analyses were performed as previously reported.1 The following antibodies were used: anti-PEAR1-EC (1/1000) (R&D), anti-histone deacetylase-1 (HDAC-1) (1/1000; H-51, Santa Cruz Biotechnology), anti-Akt-P (1/500), anti-Akt (1/1000) (Cell Signaling), anti-GFP (green fluorescent protein, 1/1000; Rockland, Gilbertsville, PA), and anti-PTEN (1/500, Heidelberg, Germany). Mature red blood cell lysates were isolated as described.15 Peripheral blood mononuclear cells were isolated via Ficoll-Paque (BD) density centrifugation, after which residual platelets and platelet-leukocyte conjugates were eliminated via magnetic cell sorting using a homemade anti-β3 monoclonal antibody to capture platelet αIIbβ3.
The use of flow cytometry and immunohistochemical staining are detailed in the online data supplement.
Zebrafish whole-mount in situ hybridization
Whole-mount RNA in situ hybridization for PEAR1 was performed with digoxigenin-labeled antisense riboprobes as previously described.16 We used as primers: forward 5′GGCGTCTCTGCGAGTGCCAG3′ and reverse 5′GCGGTGGTCACAGCGAGGAC3′. The primers used for the in situ hybridization of GATA-1 were a kind gift of Dr C. Verfaillie, Stem Cell Institute, KU Leuven.
The microinjection of morpholino antisense oligonucleotides (MOs) is detailed in supplemental Table 3.
O-Dianisidine staining of zebrafish red blood cells
Statistics analysis
Results are expressed as mean values ± standard error of the mean. Statistical significance was evaluated with unpaired Student t tests (*P < .05; **P < .01; ***P < .001).
Results
PEAR1 expression during human MK differentiation
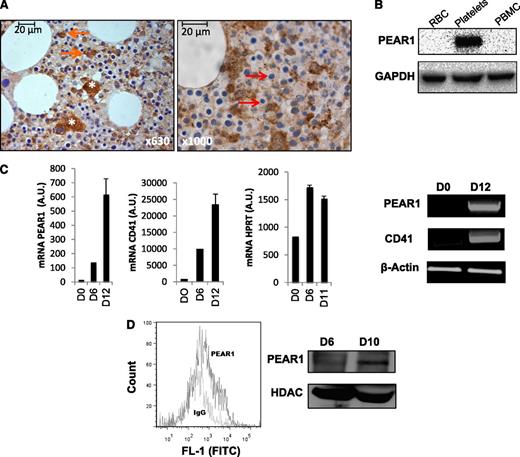
Immunohistochemical staining of normal human bone marrow showed PEAR1 positivity in MKs and revealed that granulocyte precursors, but not mature granulocytes were PEAR1 positive (Figure 1A). Also PEAR1-positive osteoblasts and endothelial cells were observed (not shown), whereas erythrocyte precursors were PEAR1 negative (Figure 1A). Western blots of isolated blood cells confirmed the presence of PEAR1 in platelets and its absence in red blood cells and peripheral blood mononuclear cells (Figure 1B). During in vitro differentiation of CD34+ cells, more detailed RT-PCR and qRT-PCR measurements showed that both PEAR1 and ITGA2B (CD41) were very low at day 0, but that their GAPDH normalized expression increased in parallel from days 6 to 12 in contrast to expression of a more stable reference, such as hypoxanthine guanine phosphoribosyl transferase (Figure 1C). Similar results were seen for both genes when tested in RT-PCR vs ACTB (β-actin) (Figure 1C). Flow cytometry and western blot analysis showed membrane PEAR1 protein expression in differentiated CD34+ cells after 6 and 10 days of maturation initiation (Figure 1D). Anti-PEAR1 antibodies triggered Akt phosphorylation in matured MKs, acting as a pseudo-ligand for PEAR1 as recently reported for platelets (not shown).1 Or, PEAR1 is progressively expressed during megakaryopoiesis and appears transiently in granulocyte precursors in the bone marrow. In the circulation, PEAR1 is only present in platelets.
PEAR1 expression in differentiating human CD34+ cells. (A) Immunohistochemical staining of PEAR1 in human bone marrow sections, at 630- and 1000-fold magnification; MKs (white asterisk), myeloid precursors (orange arrows), and erythroid precursors (red arrows) are indicated. (B) Western blot for PEAR1 in red blood cell lysates (RBC), platelets, and peripheral blood mononuclear cells (PBMC) vs GAPDH control. (C) PEAR1, ITGA2B (CD41), and hypoxanthine guanine phosphoribosyl transferase expression on day 0, day 6, and day 12 in CD34+ cells by qRT-PCR vs GAPDH control and by RT-PCR vs ACTB (β-actin) control. (D) Flow cytometric measurement of PEAR1 on the membrane in 10-day-old CD34+ cell cultures after staining with immunoglobulin G control (MFI 562) and PEAR1-Ab (MFI 1287), as indicated (representative for n = 3); western blot for PEAR1 in CD34+ cell lysates, at days 6 and 10, with HDAC-1 as loading control. 1 A.U., 1 copy for 105 copies of housekeeping gene.
PEAR1 expression in differentiating human CD34+ cells. (A) Immunohistochemical staining of PEAR1 in human bone marrow sections, at 630- and 1000-fold magnification; MKs (white asterisk), myeloid precursors (orange arrows), and erythroid precursors (red arrows) are indicated. (B) Western blot for PEAR1 in red blood cell lysates (RBC), platelets, and peripheral blood mononuclear cells (PBMC) vs GAPDH control. (C) PEAR1, ITGA2B (CD41), and hypoxanthine guanine phosphoribosyl transferase expression on day 0, day 6, and day 12 in CD34+ cells by qRT-PCR vs GAPDH control and by RT-PCR vs ACTB (β-actin) control. (D) Flow cytometric measurement of PEAR1 on the membrane in 10-day-old CD34+ cell cultures after staining with immunoglobulin G control (MFI 562) and PEAR1-Ab (MFI 1287), as indicated (representative for n = 3); western blot for PEAR1 in CD34+ cell lysates, at days 6 and 10, with HDAC-1 as loading control. 1 A.U., 1 copy for 105 copies of housekeeping gene.
PEAR1 in MK differentiation: proliferation vs maturation
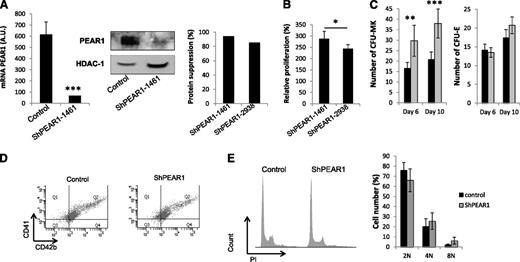
Transduction of freshly isolated human CD34+ cells with LV_miR_PEAR1_1461 decreased PEAR1 mRNA levels by 90%, compared with LV_miR_DsRed transduction (Figure 2A). Correspondingly, protein synthesis at day 10 had dropped strongly upon transduction with LV_miR_PEAR1_1461 when investigated via western blots (Figure 2A). Calculation of the degree of protein expression suppression for transduction with both viral vs the control vectors yielded up to 84% to 94% expression inhibition (Figure 2A, single experiment comparison). During CD34+ cell differentiation, proliferating cell numbers reach a plateau between days 10 and 14.19 Trypan blue counting of the cells 10 days after transduction with LV_miR_PEAR1_1461 or LV_miR_PEAR1_2938 detected >95% cell viability and uncovered 2.4- to 2.8-fold higher cell numbers of differentiating MKs following PEAR1 mRNA knockdown compared with control (P < .01 vs control; P < .05 between both vectors) (Figure 2B). More stringent clonogenic assay cultures with separate counting of CFU-MK and CFU-E colonies on days 6 and 10, confirmed that LV_miR_PEAR1_1461 transduction doubled CFU-MK colony numbers compared with control, without affecting CFU-E colony formation (Figure 2C). These findings indicate a specific role for PEAR1 in human MK progenitor proliferation, which is absent in the erythroid lineage.
PEAR1 knockdown via lentiviral vector transduction. (A) PEAR1 expression at day 10 after transduction of human CD34+ with control (LV_miR_DsRed) or LV_miR_PEAR1_1461 vector analyzed via qRT-PCR (left) and western blotting with PEAR1-EC Ab using HDAC-1 as loading control (middle). Relative suppression of the GAPDH-normalized PEAR1 protein expression at day 11 following transduction of both LV_miR_PEAR1 vectors vs control (right). (B) Relative cell proliferation at day 10 after treatment with both LV_miR_PEAR1 vectors vs control. (C) Numbers of CFU-MK and CFU-E colonies at days 6 and 10 after transduction of human CD34+ with control (black) or LV_miR_PEAR1_1461 (gray) vectors. (D) Coexpression of CD41 and CD42b during CD34+ cell differentiation analyzed by flow cytometry at day 12 after transduction of human CD34+ with control or LV_miR_PEAR1_1461 vectors, as indicated. (E) Polyploidy profiles (left) on day 10, representative of 3 experiments, after anti-CD41 labeling and DNA staining with propidium iodide (PI), after transduction of human CD34+ with control or LV_miR_PEAR1_1461 (n = 3); Ploidy histogram for control and LV_miR_PEAR1_1461 treated CD34+ cells on day 12 (n = 3). 1 A.U., 1 copy for 105 copies of housekeeping gene.
PEAR1 knockdown via lentiviral vector transduction. (A) PEAR1 expression at day 10 after transduction of human CD34+ with control (LV_miR_DsRed) or LV_miR_PEAR1_1461 vector analyzed via qRT-PCR (left) and western blotting with PEAR1-EC Ab using HDAC-1 as loading control (middle). Relative suppression of the GAPDH-normalized PEAR1 protein expression at day 11 following transduction of both LV_miR_PEAR1 vectors vs control (right). (B) Relative cell proliferation at day 10 after treatment with both LV_miR_PEAR1 vectors vs control. (C) Numbers of CFU-MK and CFU-E colonies at days 6 and 10 after transduction of human CD34+ with control (black) or LV_miR_PEAR1_1461 (gray) vectors. (D) Coexpression of CD41 and CD42b during CD34+ cell differentiation analyzed by flow cytometry at day 12 after transduction of human CD34+ with control or LV_miR_PEAR1_1461 vectors, as indicated. (E) Polyploidy profiles (left) on day 10, representative of 3 experiments, after anti-CD41 labeling and DNA staining with propidium iodide (PI), after transduction of human CD34+ with control or LV_miR_PEAR1_1461 (n = 3); Ploidy histogram for control and LV_miR_PEAR1_1461 treated CD34+ cells on day 12 (n = 3). 1 A.U., 1 copy for 105 copies of housekeeping gene.
To further delineate how PEAR1 knockdown would affect MK maturation, first, cultured cells were analyzed at day 10 for expression of the typical MK and platelet markers CD41 and CD42b as well as for their ploidy. Figure 2D shows a fractional shift toward positivity at day 10 for both markers and shows that in addition to megakaryoblasts a more mature cell subset was present, strongly positive for CD41 and CD42b. The proportion of all CD41+CD42+ positive cells at day 10 was not modified by the LV_miR_PEAR1 vector treatment (79% vs 77%, Figure 2D). Also the percentages of strongly positive CD41+CD42+ subpopulations were not different (15.7% vs 14%, Figure 2D, upper right population in Q2). Correspondingly, no major effect was observed on the histogram during propidium iodide staining of the matured cells, suggesting the mature subset to largely be the 4N MK precursor population (Figure 2E). A non-significant trend for a higher ploidy was observed for the 8N fraction in the absence of PEAR1. Therefore, to specifically investigate the proliferation rate in the late differentiation phase, LV_miR_PEAR1_1461–treated CD34+ cell cultures were incubated on day 10 with the fluorescently labeled BrdU analog EDU. The CD61 positive cell fraction in these cultures varied from 4% to 24% in different cultures; paired comparisons revealed identical CD61 positivity for LV_miR_PEAR1_1461 vs control treatment, with a mean ratio of 1.08 ± 0.13. EDU incorporation in the CD61 positive cell fraction varied from 28% to 42%, but paired comparison revealed no difference, with a mean ratio of 1.06 ± 0.045. Collectively, these results illustrate that PEAR1 primarily controls progenitor cell proliferation and not the later stages of MK maturation.
Gene (de)regulation by PEAR1 knockdown in MK progenitors
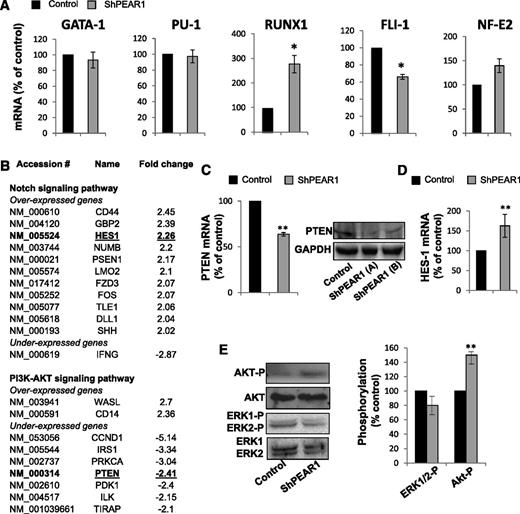
Several transcription factors have been implicated in megakaryopoiesis.20,21 Therefore, we measured the expression of GATA-1, PU-1, RUNX1, FLI-1, and NF-E2 during MK maturation in CD34+ cells (supplemental Figure 1). The expression of GATA-1 and NF-E2 increased progressively until day 12. In contrast, RUNX1 had already dropped on day 6. PU-1 expression dropped and FLI-1 mRNA levels rose during the late differentiation (n = 3, at least). Comparison of the expression levels at day 12 showed that PEAR1 depletion did not affect the expression of GATA-1 and PU-1 (Figure 3A) and had a mild effect on the expression of FLI-1 and NF-E2. In contrast, the drop in RUNX1 expression was tampered with following PEAR1 knockdown, preserving threefold higher RUNX1 mRNA levels (Figure 3A), ie, RUNX1 drops more in control than in LV_miR_PEAR1_1461–treated cells.
Transcriptional regulation and PI3K-Akt/Notch pathways after PEAR1 knockdown. (A) Proportional mRNA expression of the indicated transcription factors on day 11 in CD34+ cells transduced with control or LV_miR_PEAR1_1461 vectors vs GAPDH. (B) List of genes involved in the Notch and PI3K/Akt pathways under- or overexpressed in RT2 profiler analysis in the shPEAR1 knockdown, expressed in fold change. (C) qRT-PCR for PTEN mRNA in CD34+ cells 10 days after transduction with control and LV_miR_PEAR1_1461, relative to GAPDH; western blot for PTEN at day 10 after transduction with control and LV_miR_PEAR1_2938, relative to GAPDH in 2 separate analyses (A-B). (D) qRT-PCR for HES1 mRNA in CD34+ cells at day 10 after transduction with control and LV_miR_PEAR1_1461, relative to GAPDH. (E) Western blots for AKT-P, Akt, ERK1/2-P, and ERK1/2 in control and LV_miR_PEAR1_1461 treated CD34+ cells at day 10. Phosphorylation was quantified with ImageJ and is shown in the right histogram.
Transcriptional regulation and PI3K-Akt/Notch pathways after PEAR1 knockdown. (A) Proportional mRNA expression of the indicated transcription factors on day 11 in CD34+ cells transduced with control or LV_miR_PEAR1_1461 vectors vs GAPDH. (B) List of genes involved in the Notch and PI3K/Akt pathways under- or overexpressed in RT2 profiler analysis in the shPEAR1 knockdown, expressed in fold change. (C) qRT-PCR for PTEN mRNA in CD34+ cells 10 days after transduction with control and LV_miR_PEAR1_1461, relative to GAPDH; western blot for PTEN at day 10 after transduction with control and LV_miR_PEAR1_2938, relative to GAPDH in 2 separate analyses (A-B). (D) qRT-PCR for HES1 mRNA in CD34+ cells at day 10 after transduction with control and LV_miR_PEAR1_1461, relative to GAPDH. (E) Western blots for AKT-P, Akt, ERK1/2-P, and ERK1/2 in control and LV_miR_PEAR1_1461 treated CD34+ cells at day 10. Phosphorylation was quantified with ImageJ and is shown in the right histogram.
To characterize PEAR1 target genes in MKs grown from cytapheresis isolates, we compared control and LV_miR_PEAR1_1461–transduced CD34+ cell cultures on day 12 by profiler qRT-PCR analysis. The Notch and the PI3K/Akt pathway have been coupled to PEAR1 signaling during myeloid progenitor proliferation in mouse bone marrow cells and in platelet activation, respectively. We therefore analyzed a series of genes involved in both pathways, using RT2 profiler PCR arrays, analyzing 84 genes in each case (supplemental Figure 2).
The signal from 12 probe sets in the Notch profiler and 9 probe sets in the PI3K/Akt pathway were significantly altered following PEAR1 knockdown in differentiated CD34+ cells. Genes over- and underexpressed in the PEAR1 depleted cells vs control cells are listed according to the fold change (Figure 3B). Interestingly, we found that a target of the Notch pathway, HES1, was upregulated without expression modification of the Notch receptors (supplemental Figure 2). PTEN, a phosphatase regulating the PI3K pathway, and CCND1 (cyclin D1) were both downregulated (Figure 3B). Recently, 2 different authors described how HES1, a target of the Notch pathway, negatively regulates the transcription of PTEN during megakaryopoiesis and in mouse thymocytes.22,23 Therefore, we chose to confirm such regulation using a separate set of primers (supplemental Table 1). Figure 3C-D shows for this set of primers that PEAR1 knockdown upregulated the Notch target HES1 in parallel with the downregulation of PTEN.
Western blots for PTEN confirmed lower protein expression for the PEAR1 knockdown, both in LV_miR_PEAR1_1461– and LV_miR_PEAR1_2938–treated CD34+ cells (Figure 3C). Because PTEN regulates the PI3K/Akt pathway by dephosphorylating PIP3 into PIP2, we therefore analyzed the phosphorylation of Akt in LV_miR_PEAR1_1461–treated CD34+ cells. At day 10, the PEAR1 knockdown displayed a more intense Akt-P Ser473 band (Figure 3E), in agreement with lower PTEN activity in LV_miR_PEAR1–treated CD34+ cells, which is responsible for diminished Akt dephosphorylation. The absence of PEAR1 did not affect TPO signaling because phosphorylation of ERK1/2, a target of the c-mpl/TPO pathway, was not altered (Figure 3E).
PEAR1 silencing in zebrafish boosts thrombocyte production
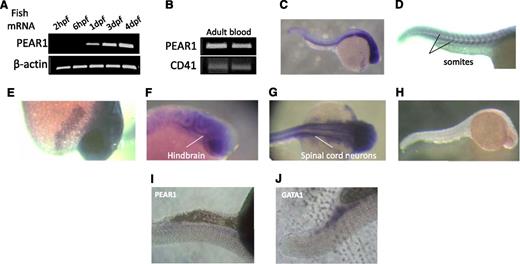
First, Pear1 expression was studied by semi-qRT-PCR during the early development of zebrafish. Pear1 mRNA appeared at 24 hours postfertilization (hpf), progressively rising until day 4 (Figure 4A). It was also detected in adult zebrafish blood (Figure 4B) and was abundantly present in the nervous system (Figure 4C,F-G) in somites (Figure 4D) and in a putative yolk sac gland (Figure 4E), as demonstrated by in situ staining for Pear1 mRNA. Staining with the sense probe was negative (Figure 4H), but Pear1 mRNA was barely detected in the caudal region (Figure 4I). To confirm this region to be the site where hematopoiesis (erythropoiesis) occurs, in situ hybridization for Gata-1 was performed (Figure 4J).
Pear1 expression in the zebrafish. (A) mRNA PEAR1 expression during fish development measured by RT-PCR (dpf, days postfertilization). (B) RT-PCR of Pear1 and CD41 in adult blood in 2 different samples. (C) Whole-mount in situ hybridization (WISH) of Pear1 at 24 hpf with a Pear1 anti-sense probe. (D) Expression in somites. (E) Expression in a putative yolk sac gland. Expression in the nervous system: midbrain-hindbrain boundary (F) and spinal cord neurons (G). (H) Control WISH of Pear1 at 24 hpf with a Pear1 sense probe. Faint Pear1 (I) and clear Gata-1 (J) expression in the caudal region.
Pear1 expression in the zebrafish. (A) mRNA PEAR1 expression during fish development measured by RT-PCR (dpf, days postfertilization). (B) RT-PCR of Pear1 and CD41 in adult blood in 2 different samples. (C) Whole-mount in situ hybridization (WISH) of Pear1 at 24 hpf with a Pear1 anti-sense probe. (D) Expression in somites. (E) Expression in a putative yolk sac gland. Expression in the nervous system: midbrain-hindbrain boundary (F) and spinal cord neurons (G). (H) Control WISH of Pear1 at 24 hpf with a Pear1 sense probe. Faint Pear1 (I) and clear Gata-1 (J) expression in the caudal region.
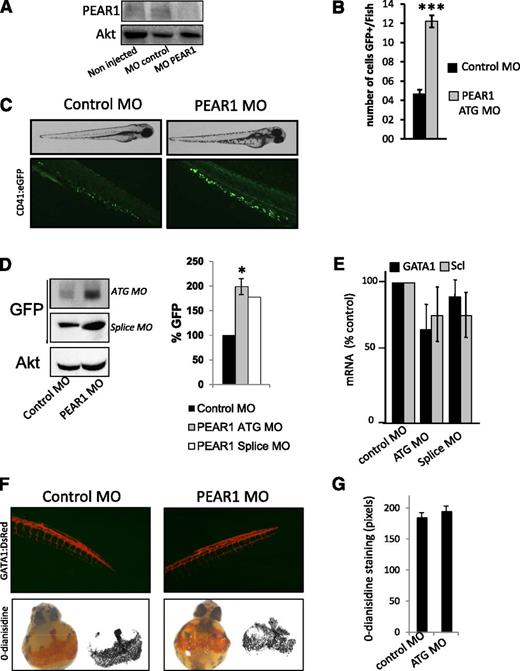
Next, we investigated the function of PEAR1 in CD41:eGFP zebrafish, which express GFP under control of the CD41 promoter. First, the potency of the PEAR1 MOs was tested on Pear1 gene expression in an in vitro luciferase assay and by RT-PCR (supplemental Methods) and on Pear1 protein expression in fish. The PEAR1 ATG MO dose-dependently inhibited the luciferase activity and the splice PEAR1 MO inhibited the PEAR1 amplification (supplemental Figure 3A-B) as well as the appearance of Pear1 in 3-day-old fish (Figure 5A). Therefore, the role of Pear1 was determined during fish development and thrombocyte production. Injection of PEAR1 MO affected the development of the zebrafish in a dose-dependent manner (supplemental Figure 3C). At low concentrations (up to 50 μM), no effects were observed. However, at a concentration equivalent to 100 μM, 42% of the fish presented a mild (23%) to severe (19%) phenotype, whereas treatment with a control MO had no effect on fish development. Defective development of the nervous system was observed (absence of eyes and head). Increasing concentrations of PEAR1 MO lead to severe phenotypic abnormalities (supplemental Figure 3C), lethality at 500 μM (not shown), and motility limitations, reflected by a higher percentage of zebrafish residing inside the chorion after 3 days compared with control zebrafish (supplemental Figure 3D). A low concentration of PEAR1 MO (50 μM) triggered an increase in the number of circulating GFP+ hematopoietic cells at day 2 (Figure 5B). Because expression of CD41 is detectable at 48 hpf in hematopoietic cells,24 the strong GFP staining (Figure 5C) further suggested these cells to be mature thrombocytes.24 Western blots confirmed elevated GFP expression in the fish (Figure 5D). To study the effect of Pear1 depletion on the development of HSCs and erythrocytes, qRT-PCR was carried out for Scl and Gata-1 on 24-hpf embryos. No expression differences were observed for these lineage markers, with only a partial specificity in zebrafish, compared with human HSC (Figure 5E).
Pear1 in zebrafish is a negative regulator of thrombocyte production. (A) Western blot for PEAR1 (PEAR1-EC Ab) in zebrafish, noninjected, injected with control MO, or PEAR1 ATG MO 3 days postfertilization (dpf); total Akt served as loading control. (B) Analysis of the number of GFP-positive cells in the tail at 2 dpf after PEAR1 MO injection vs control. (C) PEAR1 knockdown and thrombocyte production. Photography of CD41:eGFP fish at 3 dpf after injection with control or PEAR1 ATG MO with white light (top) and with fluorescent light (bottom). (D) Western blot of GFP expression and quantification at 3 dpf in whole fish after injection with control or PEAR1 ATG or PEAR1 splice MO, with Akt as loading control. (E) mRNA expression of Gata-1 and Scl, general markers for erythrocytes and HSCs, by qRT-PCR after treatment with control or PEAR1 MOs. (F) PEAR1 knockdown in erythrocyte production: photography of GATA1:dsred fish at 3 dpf after injection with control or PEAR1 ATG MO with fluorescent light (top); o-dianisidine staining of CD41:eGFP whole fish at 48 hpf and after injection with control or PEAR1 ATG MO (lower). (G) Analysis of area density for stained red blood cells after injection with control or PEAR1 ATG MO.
Pear1 in zebrafish is a negative regulator of thrombocyte production. (A) Western blot for PEAR1 (PEAR1-EC Ab) in zebrafish, noninjected, injected with control MO, or PEAR1 ATG MO 3 days postfertilization (dpf); total Akt served as loading control. (B) Analysis of the number of GFP-positive cells in the tail at 2 dpf after PEAR1 MO injection vs control. (C) PEAR1 knockdown and thrombocyte production. Photography of CD41:eGFP fish at 3 dpf after injection with control or PEAR1 ATG MO with white light (top) and with fluorescent light (bottom). (D) Western blot of GFP expression and quantification at 3 dpf in whole fish after injection with control or PEAR1 ATG or PEAR1 splice MO, with Akt as loading control. (E) mRNA expression of Gata-1 and Scl, general markers for erythrocytes and HSCs, by qRT-PCR after treatment with control or PEAR1 MOs. (F) PEAR1 knockdown in erythrocyte production: photography of GATA1:dsred fish at 3 dpf after injection with control or PEAR1 ATG MO with fluorescent light (top); o-dianisidine staining of CD41:eGFP whole fish at 48 hpf and after injection with control or PEAR1 ATG MO (lower). (G) Analysis of area density for stained red blood cells after injection with control or PEAR1 ATG MO.
At higher MO concentrations, the thrombocyte count dropped, which was related to developmental problems (supplemental Figure 3E). Correspondingly, when GATA1:dsred fish embryos were injected with the PEAR1 MO (50 μM), no differences were observed in the development of red blood cells (Figure 5F). Erythrocyte cell maturation was also studied by direct staining of red blood cells in the zebrafish yolk sac. O-dianisidine staining of whole fish embryos at 48 hpf stained red blood cells equally in control and PEAR1 MO injected embryos (Figure 5F-G). Together, these results demonstrate that mild PEAR1 suppression, compatible with normal development of zebrafish suffices to raise thrombocyte, but not erythrocyte production.
PEAR1 transcriptionally regulates PTEN, modulating Akt phosphorylation
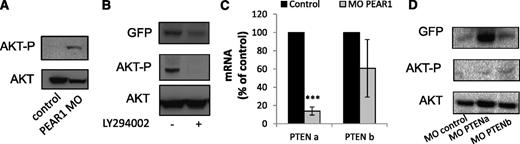
PEAR1 activates PI3K and triggers Akt phosphorylation in platelets.1 Both PI3K and the phosphatase PTEN are necessary for platelet formation, via controlling Akt phosphorylation.11 Therefore, Akt phosphorylation was investigated in the zebrafish Pear1 knockdown 3 days postfertilization after injection of PEAR1 MO or control MO. Pear1 depletion induced elevated Akt phosphorylation in total zebrafish extracts (Figure 6A). The importance of PI3K in this process is illustrated by the addition of the PI3K inhibitor LY294002, which was added to the water 30 hpf. Added at this time point, LY294002 was not lethal, but did disrupt Akt phosphorylation and thrombocyte formation (Figure 6B and supplemental Figure 4). Similar to the effect of PEAR1 neutralization in CD34+ cells during MK differentiation in vitro (Figure 3), Pten was downregulated in the Pear1 knockdown. Because the zebrafish genome encodes 2 Pten genes Ptena and Ptenb, expression of both was analyzed by qRT-PCR after PEAR1 MO injection. Ptena was potently downregulated and Ptenb weakly (Figure 6C). Therefore, the role of each phosphatase was analyzed separately in thrombocyte production and Akt phosphorylation. In agreement with earlier findings,25 the inactivation of only a single isoform did not affect fish development at 3 days postfertilization (supplemental Figure 3B). Also, both the injection of PTENa or PTENb MO increased Akt phosphorylation, even though the thrombocyte count was only increased after PTENa MO treatment, as shown by the GFP western blots (Figure 6D). Combined, these results demonstrate that Pear1 controls Ptena expression, a phosphatase regulating thrombocyte formation, via Akt dephosphorylation.
Pear1 controls Ptena, a regulator of thrombocyte production. (A) Western blot of Akt-P (Ser473) at 3 days postfertilization (dpf) in zebrafish after injection of control or PEAR1 MO. (B) Western blot for GFP and Akt-P in whole fish after treatment with LY294002 or DMSO, added at 30 hpf and analyzed at 3 dpf, with Akt as a loading control. (C) mRNA expression of Ptena and Ptenb by qRT-PCR 3 dpf after injection of control or PEAR1 MO, expressed as percentage of control (n = 3). (D) Western blot analysis of GFP, Akt-P, and Akt after injection of PTENa or PTENb MO 3 dpf after injection.
Pear1 controls Ptena, a regulator of thrombocyte production. (A) Western blot of Akt-P (Ser473) at 3 days postfertilization (dpf) in zebrafish after injection of control or PEAR1 MO. (B) Western blot for GFP and Akt-P in whole fish after treatment with LY294002 or DMSO, added at 30 hpf and analyzed at 3 dpf, with Akt as a loading control. (C) mRNA expression of Ptena and Ptenb by qRT-PCR 3 dpf after injection of control or PEAR1 MO, expressed as percentage of control (n = 3). (D) Western blot analysis of GFP, Akt-P, and Akt after injection of PTENa or PTENb MO 3 dpf after injection.
Discussion
This study reports that PEAR1 is progressively upregulated during human megakaryopoiesis and that it is a negative regulator of MK progenitor cell proliferation, but not of MK maturation. These observations were confirmed in zebrafish, where knockdown of PEAR1 enhances thrombopoiesis but not erythropoiesis. The expression of various genes, implicated in the PI3K/Akt and Notch pathways as well as in gene transcription, was mildly to moderately modified in the PEAR1 knockdowns. PTEN, a phosphatase that regulates the degree of Akt phosphorylation during megakaryopoiesis, was also downregulated. Hence, PEAR1 elimination causes chronically elevated Akt-P levels, thus enhancing MK progenitor cell proliferation.
The differentiation of cultured CD34+ cells is accompanied by PEAR1 upregulation until day 12, coinciding with the expression of CD41 and GATA-1. The parallelism in the expression profiles of PEAR1 and GATA1 is in line with the presence of GATA-1 binding sites in the 5′-upstream sequence of the PEAR1 promoter, identified via the TFMATRIX transcription factor binding site profile database (http://www.cbrc.jp/research/db/TFSEARCH.html) and as reported by Farnham et al.26 Whereas progressive upregulation of nuclear factor E2 is not surprising for a transcription factor implicated in the late stages of MK maturation and proplatelet formation,27 the drop in RUNX1 expression was unexpected. RUNX1 regulates constituents of the MK and platelet cytoskeleton during late megakaryopoiesis and platelet formation27 and shows a functional cooperation with GATA-1.28 The earlier demonstration that depletion of RUNX1 in UT-7/GM cells caused upregulation of MK markers and also polyploidization, accompanied by reduced cell proliferation,29 is in line with the present findings. Correspondingly, the mildly weaker drop of RUNX1 in shPEAR1-treated CD34+ cells may reflect the higher proliferation of MK progenitors during the first few days of culture.
Overexpression of Pear1 in mouse bone marrow cells reduces myeloid progenitor proliferation. Moreover, the ectopic expression of Pear1 in NIH 3T3 fibroblasts reduced both early and late myeloid progenitors in nonadherent cocultured bone marrow cells.3 It is not clear whether this loss is caused by apoptosis or artificial signaling coupled to excessive PEAR phosphorylation.1 We therefore investigated the role of PEAR1 in megakaryopoiesis by reducing its expression in 2 distinct models based on expression depletion. In human CD34+ cells, PEAR1 was reduced, employing lentiviral vectors encoding different miRNA-based short hairpins (LV_miR_PEAR1_1461 and LV_miR_PEAR1_2938); in zebrafish embryos, it was counteracted via injection of 2 types of morpholino. Both approaches demonstrated that PEAR1 negatively regulates the early steps of megakaryopoiesis and thrombopoiesis, respectively.
Several approaches distinguishing MK progenitor proliferation and MK maturation pointed toward a role for PEAR1 in proliferation primarily. This is illustrated by the gradual expression of CD41 in megakaryoblasts and by the similar distribution of CD41 over smaller and larger differentiating MK precursors after control or shPEAR1 treatment. Zebrafish CD41 positivity has been demonstrated as early as 42 hpf and it appears at 48 hpf in circulating hematopoietic cells. Because CD41-GFP zebrafish allow the identification of mature thrombocytes with high GFP positivity24 vs their precursors,30 this model was suited to studying Pear1 neutralization in thrombopoiesis. The model also illustrated that the rapid stimulation of thrombopoiesis was without effect on erythropoiesis. Whereas we cannot exclude that the Pear1 knockdown affects other circulating cells (excluding red blood cells) in zebrafish, in human blood, PEAR1 is exclusively expressed in circulating platelets. PEAR1 is also expressed in human myeloid precursors—the relevance of which remains to be elucidated.
Both in MK progenitors and zebrafish, a knockdown of PEAR1 affected the transcriptional regulation of several genes, including PTEN. PTEN is a tumor suppressor protein and mutations in PTEN have been observed in a variety of malignancies, including leukemia. Correspondingly, the transient silencing of PTEN in human CD34+ cells enhanced their proliferative potential and ability to engraft mice.31 The present study shows that absence of PEAR1 also causes a partial silencing of PTEN, leading to a new steady state in proliferating CD34+ cells, with elevated baseline levels of Akt-P stimulating cell proliferation. Formally, this situation differs from the ligand-induced activation of PEAR1 triggering cellular signaling, which is transient and short, via reversible Akt phosphorylation.1
Pten+/− mice exhibit a predisposition to a variety of malignancies. In man, it remains to be investigated whether PEAR1 mutations exist in oncogenic tissues where PEAR1 is expressed and whether a link exists with PTEN expression. However, several cases of breast, prostate, endometrial, and liver cancers exhibited weak to moderate cytoplasmic immunoreactivity for PEAR1 (http://www.proteinatlas.org/ENSG00000187800/cancer).
The zebrafish genome encodes 2 Pten genes: Ptena and Ptenb. Both have similar lipid phosphatase activity.25 In the Pear1 knockdown, we observed an association between elevated thrombocyte formation and downregulated Ptena, but not Ptenb. The MO-mediated knockdown of Ptena, but not Ptenb, resulted in upregulated thrombopoiesis, despite a similar increase in Akt phosphorylation in both of them. Neither PTENa nor PTENb morpholinos affected zebrafish viability, development, and fertility, which is in agreement with similar findings in homozygous single mutant zebrafish lacking Ptena or Ptenb.25 Hence, even when embryonic development is not affected by the PTENa MO, megakaryopoiesis and thrombocyte formation depend on Pear1, at least in part, via the regulation of Ptena. Recent studies in MKs22 and in thymocytes23 have revealed that Hes1 is a negative regulator of Pten expression. We presently found evidence of HES1 upregulation in the PEAR1 knockdown in CD34+ cells, but not in zebrafish. The regulation of PTEN expression is still unclear. Recent studies have shown that PTEN may both be positively and negatively regulated transcriptionally as well as posttranslationally by phosphorylation, oxidation, and acetylation. Transcription factors known to be involved include peroxisome proliferator-activated receptor γ,32 EGR1,33 and p53.33 Several factors negatively regulate PTEN, including miRNAs,34 17β-estradiol,35 MKK4,36 JUN,37 IGF-1,38 and transforming growth factor-β.39 Furthermore, the expression of PTEN appears to be dependent also on nuclear factor κB,35 linking MK differentiation and apoptosis.40 Western blots confirmed that reduced mRNA levels in CD34+ cells coincided with weaker PTEN protein bands.
Src family tyrosine kinases (SFKs) have been identified as negative regulators of thrombopoiesis. A reduced PEAR1 expression inevitably interferes with PEAR1-dependent SFK signaling. Hence, our present findings that the loss of PEAR1 increases thrombopoiesis comply with these observations that SFK inhibitors and mice deficient in Lyn enhance MK proliferation, maturation, polyploidy, and platelet release in cell culture.41,42 Therefore, PEAR1 may join other previously described negative regulators such as SFKs, focal adhesion kinase, platelet factor 4, thrombospondin-1,43 and the pituitary adenylyl cyclase-activating peptide.44
In addition to the role of PEAR1 in platelet function,1 megakaryopoiesis, and thrombocyte formation (this study), PEAR1 is abundantly present in the neuronal crest of zebrafish where it appears to be necessary for embryo development. Progressively increasing PEAR1 MO concentrations caused a variety of central nervous system defects. Previously, Wu et al45 identified PEAR1 in glial precursor cells as an engulfment receptor implicated in the phagocytosis of dead sensory neurons. The developmental problem in PEAR1 knockdowns is therefore compatible with this putative role for PEAR1 in the clearance of the sensory neuron corpse. Thus, a defective phagocytosis in PEAR1 MO treated zebrafish embryos may very well be the cause of the developmental phenotypes observed, further illustrating the importance of neuronal crest PEAR1.
In conclusion, the present study shows that a reduced PEAR1 expression in MK progenitors lowers Akt dephosphorylation secondarily to transcriptional downregulation of PTEN, an intervention promoting MK progenitor proliferation and indirectly enhancing megakaryopoiesis and thrombopoiesis.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors appreciate the skillful assistance of Serena Loyen, Katrien Cludts, Els Audenaarde, and Christine Wittevrongel.
This study was supported by research grant Krediet aan Navorsers 1.5.229.11N from the Fonds Weternschappelijk Onderzoek (FWO) Vlaanderen and by FWO Vlaanderen grant G.0601.12N. The Center for Molecular and Vascular Biology is supported by the Programmafinanciering KU Leuven (PF/10/014) and the Geconcerteerde Onderzoeksacties (GOA 2009/13) from the University of Leuven.
Authorship
Contribution: A.K. designed and performed research, analyzed data, and wrote the manuscript; C.V. and T.T. performed research; S.L. performed zebrafish experiments and advised on zebrafish research methodology; K.F. advised on MK/platelet research methodology and performed clonogenic assays; R.G. made lentiviral vectors; P.V. and M.F.H. designed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marc Hoylaerts, Center for Molecular and Vascular Biology (CMVB), Department of Cardiovascular Sciences, University of Leuven, Herestraat 49, B3000 Leuven, Belgium; e-mail: Marc.Hoylaerts@med.kuleuven.be.
References
Author notes
A.K. and C.V. contributed equally to the study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal