Key Points
The efficacy and safety of a novel redirected T-cell–based adoptive immunotherapy targeting hTERT for patients with adult T-cell leukemia.
hTERT-specific T-cell receptor gene-transduced CD8+ T cells lyse ATL cells, but not normal cells, both in vitro and in vivo.
Abstract
Although adult T-cell leukemia (ATL) has a poor prognosis, successful allogeneic hematopoietic stem cell transplantation (allo-HSCT) in some cases suggests that a cellular immune-mediated strategy can be effective. So far, however, no effective target for anti-ATL immunotherapy has been defined. Here we demonstrated for the first time that human telomerase reverse transcriptase (hTERT) is a promising therapeutic target for ATL, and we developed a novel redirected T-cell–based immunotherapy targeting hTERT. hTERT messenger RNA was produced abundantly in ATL tumor cells but not in steady-state normal cells. Rearranged human leukocyte antigen-A*24:02 (HLA-A*24:02) –restricted and hTERT461-469 nonameric peptide-specific T-cell receptor (TCR) α/β genes were cloned from our previously established cytotoxic T lymphocyte clone (K3-1) and inserted into a novel retroviral TCR expression vector encoding small interfering RNAs for endogenous TCR genes in redirected T cells (hTERT-siTCR vector). Consequently, allogeneic or autologous gene-modified CD8+ T cells prepared using the hTERT-siTCR vector successfully killed ATL tumor cells, but not normal cells including steady-state hematopoietic progenitors, in an HLA-A*24:02-restricted manner both in vitro and in vivo. Our experimental observations support the development of a novel hTERT-targeting redirected T-cell–based adoptive immunotherapy for ATL patients, especially those for whom suitable allo-HSCT donors are lacking.
Introduction
Adult T-cell leukemia (ATL) is an aggressive peripheral T-cell neoplasm caused by human T-cell lymphotropic virus I (HTLV-I).1 It is estimated that there are more than 1 million HTLV-I carriers in Japan, about 5% of whom develop ATL at around 60 years of age or older.2 Because ATL tumor cells soon acquire chemotherapy resistance and compromise host immunity against infectious pathogens, ATL has a poor prognosis.3 Although most ATL patients are ineligible for allogeneic hematopoietic stem cell transplantation (allo-HSCT) because of advanced age, age-related comorbidity, or lack of suitable donors,4 the number of ATL patients who are treated successfully with allo-HSCT and achieve prolonged survival has been increasing.5 The graft-versus-ATL effect observed in ATL patients treated successfully with allo-HSCT5 strongly suggests that a cellular immune-mediated approach for ATL can be clinically effective. With regard to cellular immunotherapy for ATL (unlike Epstein-Barr virus [EBV]-associated malignancy6 ), targeting of antigens associated with HTLV-I (the causative virus of ATL) such as Tax7 and HBZ8 still remains controversial, and the recently proposed NY-ESO-19 (a cancer-testis antigen) still awaits clinical validation. Thus, at this time, no effective therapeutic target antigen for anti-ATL immunotherapy has been clinically defined.
Human telomerase reverse transcriptase (hTERT), which is a component of human telomerase and a catalytic subunit for telomere elongation, is activated in almost all cancer cells, including hematologic malignancies, but not in normal cells.10 In HTLV-I–infected cells and ATL tumor cells, Tax or interleukin-2 (IL-2) signaling strongly activates the hTERT promoter through the nuclear factor-κB or PI3K pathway,11-13 suggesting that expression of hTERT protein would be upregulated in ATL tumor cells. Clinical trials of anticancer immunotherapy targeting hTERT have already been conducted, and both the safety and induction of immune responses to hTERT have been reproducibly confirmed.10,14-17 In our previous studies, we defined a [human leukocyte antigen] HLA-A*24:02-restricted hTERT461-469 nonameric peptide (VYGFVRACL) that was capable of inducing antileukemia cytotoxic T lymphocytes (CTLs),18 and we subsequently established a CTL clone, K3-1, specific for this epitope.19 We previously conducted a phase I/II clinical trial of hTERT peptide vaccine for treatment of HLA-A*24:02+ patients with lung cancer and metastatic renal cell cancer.20 These achievements strongly encouraged us to further explore cellular immune-mediated treatment of ATL targeting hTERT. Because of concern over the potential regulatory T-cell function of ATL tumor cells,21 in this study we focused on developing a redirected T-cell–based immunotherapy targeting hTERT rather than using an hTERT461-469 peptide vaccine. Recently developed forms of anticancer immunotherapy using gene-modified T cells that redirect defined tumor-associated antigens have been shown to have clinical promise.22-25 To this end, therefore, we first cloned the rearranged HLA-A*24:02-restricted and hTERT461-469-specific T-cell receptor α/β (TCR-α/β) genes from K3-1 and inserted them into a novel TCR gene expression vector carrying silencers for endogenous TCRs (siTCR vector)26 in redirected T cells (hTERT-siTCR vector). Notably, we used a souped-up second-generation 2A peptide-based siTCR vector that achieved an increased level of expression of the introduced TCR.27
In this study, we used the newly established hTERT-siTCR vector to examine the feasibility of a novel redirected T-cell–based adoptive immunotherapy targeting hTERT for treatment of ATL.
Patients and methods
Cell lines, freshly isolated leukemia cells, and normal cells
Approval for this study was obtained from the institutional review board of Ehime University Hospital. Written informed consent was obtained from all patients, healthy volunteers, and parents of cord blood donors in accordance with the Declaration of Helsinki.
B-lymphoblastoid cell lines (B-LCLs) were established by transformation of peripheral blood B lymphocytes with EBV. ATN-1,28 TL-Om1,29 HUT10229 , and TL-MAT30 were human T-cell lines established from ATL patients, and TL-Su,31 MT-1,32 MT-232 , and MT-433 were human T-cell lines transformed by HTLV-I infection. LCLs, T2-A24,19 K562 (American Type Culture Collection [ATCC]), and human T-cell lines (except TL-Om1), maintenance of which requires 10 U/mL recombinant human IL-2 (rhIL-2) (R&D Systems), were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum. The HLA-A*24:02 gene-transduced K562 (K562-A24) was maintained in culture medium supplemented with 1.0 μg/mL puromycin (Sigma-Aldrich). Peripheral blood mononuclear cells (PBMCs) from ATL patients and healthy donors and cord blood mononuclear cells (CBMCs) from healthy donors were isolated by density gradient centrifugation and stored in liquid nitrogen until use. All samples from ATL patients contained more than 90% ATL cells. CD4+ T cells, CD14+ cells from PBMCs, and CD34+ cells from CBMCs were isolated by using CD4+ cell-, CD14+ cell-, or CD34+ cell-isolating immunomagnetic beads (MACS beads; Miltenyi Biotec), respectively. IL-2–dependent CD4+ cell lines induced by HTLV-I infection were generated as reported previously.8
Cloning of full-length TCR α and β chain genes and construction of hTERT-siTCR retroviral vector
HLA-A*24:02-restricted and hTERT461-469 nonameric peptide (VYGFVRACL)-specific TCR-α/β genes were cloned from our previously established CTL clone, K3-1,19 by using the 5′ rapid amplification of complementary DNA ends method (Clontech). The rearranged TCR-α/β genes of K3-1 expressed the germ line gene segments TRAV29DV5/TRAJ34/TRAC and TRBV20-1/TRBJ2-1/TRBC2, respectively. The retroviral vector expressing K3-1–derived TCR genes was constructed as reported previously.26,27,34 Briefly, the constant regions of the hTERT-specific TCR-α/β genes were codon optimized and then integrated into a novel Splice-b2Aa-siTCR–based retroviral vector encoding small interfering RNAs that complementarily bind to the constant regions of the endogenous TCR-α/β genes (hTERT-siTCR vector).27
Establishment of hTERT-siTCR–transduced CD8+ T-cell lines
Isolated CD8+ T cells from PBMCs of healthy volunteers or ATL patients using CD8+ cell-isolating MACS beads and stimulation with 1 μg/mL anti-CD3 monoclonal antibody (mAb; OKT-3; BioLegend) were cultured in GT-T503 (Takara Bio) supplemented with 5% human serum, 0.2% human albumin, 50 U/mL rhIL-2, 5 ng/mL rhIL-7 (R&D Systems), 10 ng/mL rhIL-15 (PeproTech), and 10 ng/mL rhIL-21 (Shenandoah Biotechnology). Then, CD8+ T cells were transfected with the hTERT-siTCR retroviral vector using RetroNectin (Takara Bio) -coated plates as described previously.34 In some experiments, because TRBV20-1 is specifically labeled with anti-Vβ2 mAb (IMGT Web resources: http://www.imgt.org/), Vβ2-positive cells among hTERT-siTCR–transduced CD8+ T cells (hTERT-siTCR/CD8) were further isolated by using fluorescein isothiocyanate (FITC) –conjugated Vβ2 mAb (Beckman Coulter) and anti-FITC–conjugated MACS beads. To measure the expression levels of the introduced hTERT-specific TCR in gene-modified CD8+ T cells, the cells were labeled with anti-CD8 (BD Biosciences) and anti-Vβ2 mAbs and phycoerythrin-conjugated HLA-A*24:02/hTERT461-469 tetramer or HLA-A*24:02/HIV-1 Env584-592 (RYLRDQQLL) tetramer, as a negative control.19 Labeled cells were analyzed by using a Gallios flow cytometer (Beckman Coulter) and FlowJo Version 7.2.2 software (TreeStar). To expand the hTERT-siTCR/CD8 cells, they were stimulated weekly with mitomycin-C (Kyowa Hakko) –treated and hTERT461-469 peptide–pulsed HLA-A*24:02+ LCLs.
Cytotoxicity assays
Standard 51Cr-release assays were performed as described previously.35 Briefly, 5 × 103 unpulsed or peptide-pulsed target cells were labeled with 51Cr (Na251CrO4; MP Bio Japan) and incubated at various ratios with effector cells in 200 μL of culture medium in 96-well round-bottomed plates. To assess HLA class I restriction, target cells were incubated with 10 μg/μL anti-HLA class I framework mAb (clone w6/32; ATCC) or a control anti-HLA -DR mAb (clone L243; ATCC) for 1 hour, then incubated with effector cells for 5 hours. The percentage of specific lysis was calculated as (experimental release cpm – spontaneous release cpm)/(maximal release cpm – spontaneous release cpm) × 100 (%). In some experiments, time-lapse imaging was used. Ten thousand ATL cells lentivirally gene-modified to express monomeric Azami-Green (Amalgaam) were cocultivated with 5 × 104 effector cells expressing hTERT-specific TCR (at an effector:target ratio of 5:1) for 12 hours in culture medium supplemented with 10 μg/mL propidium iodide (Sigma) to label dead cells red by using a glass dish for microscopic observation of live cells (iBIDI-dish1 Hi-Q4; Nikon). Images were acquired by using a systemic bio-imaging tool (BioStation IM; Nikon). To examine the cytotoxicity of these effector cells against early-differentiated and highly proliferating subsets of hematopoietic progenitor cells, CB-CD34+ cells cultured by using a hematopoietic cell expansion medium (StemSpan CC100 and StemSpan SFEM; Stem Cell) for 7 days were subjected to flow-based cytotoxicity assay. 7-Aminoactinomycin D (7-AAD) –positive dead cells in each subset were examined by flow cytometry.
Quantitative analysis of hTERT mRNA expression
Quantitative real-time PCR (qRT-PCR) for hTERT messenger RNA (mRNA) was performed as described previously.36 Briefly, after complementary DNA was synthesized, qRT-PCR for hTERT mRNA (NM_198253) was performed by using the QuantiTect SYBR green PCR Kit (QIAGEN) and primers as follows: forward, 5′-TTCTTGTTGGTGACACCTCACCTC-3′; reverse, 5′-CAGCCATACTCAGGGACACCTC-3′ (Takara Bio). Human hypoxanthine phosphoribosyltransferase 1 (hHPRT1) mRNA (NM_000194) was prepared and used as an internal control. Samples were analyzed by using an ABI Prism 7500 Sequence Detection System (Applied Biosystems). The expression level of hTERT mRNA was corrected by reference to that of hHPRT1 mRNA, and the amount of hTERT mRNA relative to that in PBMCs was calculated by the comparative threshold cycle method. K562, which strongly expresses hTERT mRNA, was used as an internal control.
Western blotting of hTERT protein
For analysis of protein expression, western blotting was performed as described previously.35 Briefly, cell lysates were subjected to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (e-PAGEL, ATTO) and blotted onto polyvinylidene difluoride membranes (Bio-Rad Laboratories). The blots were incubated first with anti-hTERT rabbit mAb (Millipore), then with horseradish peroxidase–conjugated anti-rabbit immunoglobulin G Ab (GE Healthcare). The probed proteins were visualized by using an enhanced chemiluminescence system (GE Healthcare). Subsequently, the blotted membranes were stripped and reprobed with anti–β-actin mouse mAb (Sigma-Aldrich) to confirm equivalent protein loading between samples.
Detection of hTERT461-469–specific CTL precursors in the periphery of ATL patients
PBMCs from HLA-A*24:02+, HLA-A*24:02− ATL patients, or HLA-A*24:02+ healthy individuals were seeded in 24-well plates at 1.5 × 106 per well in the presence of the hTERT461-469 peptide at a concentration of 1 μM in GT-T503 medium supplemented with 5% human serum and 10 U/mL IL-2. After culturing for 14 days, cultured PBMCs were stained with FITC-conjugated anti-CD8 mAb and HLA-A*24:02/hTERT461-469 tetramer or control tetramer at a concentration of 20 μg/mL at 4°C for 20 minutes. Subsequently, the stained cells were analyzed by flow cytometry.
IFN-γ secretion assay
hTERT-siTCR/CD8 or K3-1 (2 × 104) cells were incubated with 2 × 104 hTERT461-469 peptide-pulsed (1 μM) or unpulsed K562-A24 or K562 cells for 24 hours. Interferon gamma (IFN-γ) in the culture supernatant was measured by using an enzyme-linked immunosorbent assay kit (Pierce). Enzyme-linked immunospot assays were used to detect the epitope-responsive IFN-γ production mediated by hTERT461-469-specific CTL precursors in the periphery of ATL patients as described previously.34
Anti-ATL tumor effect of hTERT-siTCR–transduced CD8+ T cells in xenografted mouse models
To assess the in vivo anti-ATL tumor effect mediated by hTERT-siTCR/CD8, a bioluminescence assay using a xenografted mouse model was used. First, we lentivirally generated a luciferase gene–transduced HLA-A*24:02+ ATL cell line, ATN-1 (ATN-1/luc). For measurement, anesthetized xenografted mice were given an intraperitoneal injection of 2.5 mg/body VivoGlo luciferin (Caliper Life Science), and images were acquired for 5 to 10 minutes by using an AEQUORIA luminescence imaging system (Hamamatsu Photonics). The acquired photon counts were analyzed by using AQUACOSMOS software (Hamamatsu Photonics).
Six-week-old NOD/scid/γcnull (NOG) female mice37 were purchased from the Central Institute for Experimental Animals and maintained in the institutional animal facility at Ehime University. All in vivo experiments were approved by the Ehime University animal care committee. For the Winn assay, 5 × 105 ATN-1/luc cells and 2.5 × 106 hTERT-siTCR/CD8 or non–gene-modified CD8+ T cells (NGM/CD8) were subcutaneously inoculated into the abdominal wall of NOG mice that had been pretreated with 1 Gy irradiation. Thereafter, 2.5 × 106 effector cells of each type were administered weekly to the corresponding mice, respectively, via the tail vein for a total of 3 times. For the adoptive transfer experiments, similarly pretreated mice were intravenously inoculated with 5 × 105 ATN-1/luc cells. After 4 days, mice started to receive intravenously infused 5 × 106 hTERT-siTCR/CD8 or NGM/CD8, respectively, for a total of 5 times. These mice were serially monitored for tumor growth determined by photon counts acquired every 7 days until they were euthanatized owing to disease progression.
Statistical analysis
The Mann-Whitney U test was used to assess differences between two groups; a P value of < .05 was considered significant.
Results
ATL tumor cells abundantly express hTERT mRNA and hTERT protein
The expression level of hTERT mRNA in the ATL/HTLV-I–infected cell line (n = 8), freshly isolated tumor cells from ATL patients (n = 10), normal PBMCs from healthy individuals (n = 6), and CD34+ cells from normal CBMCs (CB-CD34+) (n = 3) were measured by using the qRT-PCR method. hTERT mRNA expression relative to normal PBMCs was 21.3 ± 17.9 for the ATL/HTLV-I–infected cell line, 7.48 ± 6.89 for freshly isolated ATL tumor cells, and 1.10 ± 0.12 for CB-CD34+ cells (mean ± standard deviation). In Figure 1A, the ATL/HTLV-I–infected cell line and freshly isolated ATL tumor cells, but not CB-CD34+, abundantly produced hTERT mRNA in comparison with normal PBMCs, the difference being statistically significant. The P value was .002 for the ATL/HTLV-I–infected cell line, .001 for freshly isolated ATL tumor cells, and .243 for CB-CD34+ cells. Similarly, western blotting demonstrated abundant expression of hTERT protein in the ATL tumor cells (Figure 1B).
Abundant expression of hTERT in ATL tumor cells. (A) Expression of hTERT mRNA in ATL/HTLV-I infected cell lines (▪), freshly isolated ATL tumor cells from patients (▲), normal PBMCs (○), and CB-CD34+ cells (△) were examined by qRT-PCR. The level of hTERT mRNA expression in the K562 leukemia cell line (●) was used as an internal control. The expression level of hTERT mRNA in each sample was calculated relative to that of PBMCs. hTERT mRNA expression relative to normal PBMCs was 21.3 ± 17.9 for the ATL/HTLV-I–infected cell line, 7.48 ± 6.89 for freshly isolated ATL tumor cells, and 1.10 ± 0.12 for CB-CD34+ (mean ± standard deviation [SD]). The ATL/HTLV-I–infected cell line and freshly isolated ATL tumor cells expressed hTERT mRNA abundantly and significantly (*P < .01). (B) Expression of hTERT protein in ATL cell lines and normal PBMCs was confirmed by western blotting.
Abundant expression of hTERT in ATL tumor cells. (A) Expression of hTERT mRNA in ATL/HTLV-I infected cell lines (▪), freshly isolated ATL tumor cells from patients (▲), normal PBMCs (○), and CB-CD34+ cells (△) were examined by qRT-PCR. The level of hTERT mRNA expression in the K562 leukemia cell line (●) was used as an internal control. The expression level of hTERT mRNA in each sample was calculated relative to that of PBMCs. hTERT mRNA expression relative to normal PBMCs was 21.3 ± 17.9 for the ATL/HTLV-I–infected cell line, 7.48 ± 6.89 for freshly isolated ATL tumor cells, and 1.10 ± 0.12 for CB-CD34+ (mean ± standard deviation [SD]). The ATL/HTLV-I–infected cell line and freshly isolated ATL tumor cells expressed hTERT mRNA abundantly and significantly (*P < .01). (B) Expression of hTERT protein in ATL cell lines and normal PBMCs was confirmed by western blotting.
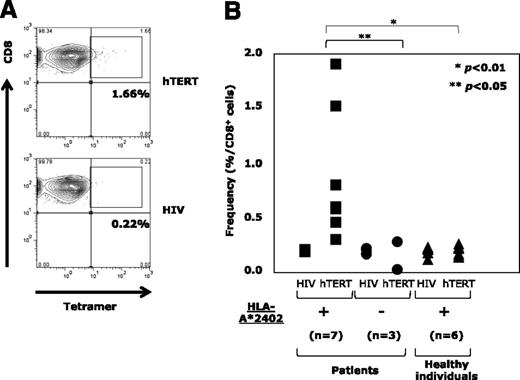
Circulatory hTERT461-469-specific CTL precursors were exclusively detectable in the periphery of HLA-A*24:02+ ATL patients
Next, by using the tetramer assay, we examined circulatory hTERT461-469-specific CTL precursors in PBMCs from HLA-A*24:02+ ATL patients (n = 7), HLA-A*24:02− ATL patients (n = 3) before chemotherapy, and HLA-A*24:02+ healthy individuals as controls (n = 6). Since freshly isolated PB lymphocytes were almost negative for tetramer staining, PBMCs stimulated with hTERT461-469 peptide were analyzed. A representative example of an HLA-A*24:02+ ATL patient is shown in Figure 2A. The frequencies of hTERT461-469-specific CTL precursors in HLA-A*24:02+ and HLA-A*24:02− ATL patients and HLA-A*24:02+ healthy individuals are summarized in Figure 2B. hTERT461-469-specific CTL precursors were detected at 0.88% ± 0.55% in HLA-A*24:02+ ATL patients, being significantly more frequent than in HLA-A*24:02− ATL patients (0.11% ± 0.1%; P < .05) or HLA-A*24:02+ healthy individuals (0.2% ± 0.04%; P < .01). These observations confirmed the presence of primed memory CD8+ T cells with hTERT461-469 epitope/HLA-A*24:02 complex (ie, that the hTERT461-469 epitope must be naturally immunogenic) in HLA-A*24:02+ ATL patients.
Detection of circulatory hTERT461-469-specific CTL precursors in the periphery of ATL patients. (A) hTERT461-469-specific CTL precursors in PBMCs repetitively stimulated with hTERT461-469 peptide from HLA-A*24:02+ ATL patients were detected by using HLA-A*24:02/hTERT461-469 tetramer. A representative case is shown. HLA-A*24:02/HIV tetramer was used as a negative control. (B) In comparison with HLA-A*24:02− ATL patients (●) (n = 3) and HLA-A*24:02+ healthy individuals (▲) (n = 6), the frequency of hTERT461-469-specific CTL precursors in HLA-A*24:02+ ATL patients (▪) (n = 7) was significantly high (*P < .01; **P < .05). The frequency was 0.88% ± 0.55% for HLA-A*24:02+ ATL patients, 0.11% ± 0.1% for HLA-A*24:02− ATL patients, and 0.2% ± 0.04% for HLA-A*24:02+ healthy individuals (mean ± SD).
Detection of circulatory hTERT461-469-specific CTL precursors in the periphery of ATL patients. (A) hTERT461-469-specific CTL precursors in PBMCs repetitively stimulated with hTERT461-469 peptide from HLA-A*24:02+ ATL patients were detected by using HLA-A*24:02/hTERT461-469 tetramer. A representative case is shown. HLA-A*24:02/HIV tetramer was used as a negative control. (B) In comparison with HLA-A*24:02− ATL patients (●) (n = 3) and HLA-A*24:02+ healthy individuals (▲) (n = 6), the frequency of hTERT461-469-specific CTL precursors in HLA-A*24:02+ ATL patients (▪) (n = 7) was significantly high (*P < .01; **P < .05). The frequency was 0.88% ± 0.55% for HLA-A*24:02+ ATL patients, 0.11% ± 0.1% for HLA-A*24:02− ATL patients, and 0.2% ± 0.04% for HLA-A*24:02+ healthy individuals (mean ± SD).
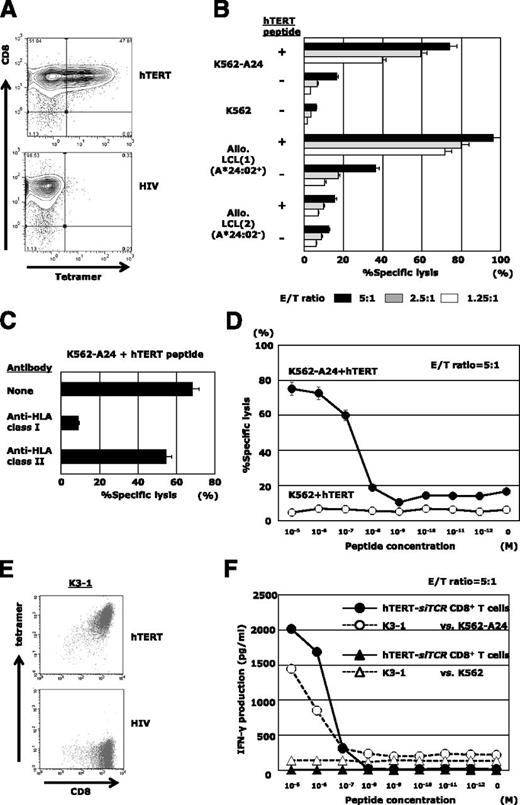
hTERT-siTCR–transduced CD8+ T cells exert anti-ATL reactivity in vitro
The hTERT-siTCR gene was retrovirally introduced into normal CD8+ T cells. Transduction efficiency determined by expression of Vβ2 on the gene-modified T cells was 85% to 95% (data not shown), and almost 50% of the transfectants were positive for HLA-A*24:02/hTERT461-469 tetramer (Figure 3A). The cognate epitope specificity and HLA-A*24:02 restriction were examined by using standard 51Cr-release assays (Figure 3B). Because expression of hTERT mRNA in LCLs was upregulated (supplemental Figure 2C), hTERT peptide-unpulsed HLA-A*24:02+ LCLs were killed to some extent, reflecting the presence of endogenously processed hTERT (Figure 3B). Such epitope-specific cytotoxicity mediated by hTERT-siTCR/CD8 was obviously attenuated by anti-HLA class I mAb, but not by anti-HLA-DR mAb (Figure 3C). The antigen sensitivity to cognate hTERT461-469 peptide mediated by hTERT-siTCR/CD8 (shown in Figure 3D) was similar to that of the parental CTL clone, K3-1 (Figure 3E-F).
hTERT-siTCR–transduced CD8+ T cells display epitope-specific responsiveness. (A) Representative flow cytometry plots showing staining of hTERT-siTCR–transduced CD8+ T cells with HLA-A*24:02/hTERT461-469 tetramer. HLA-A*24:02/HIV tetramer was used as a negative control. (B) 51Cr-release assays were conducted by using hTERT-siTCR–transduced CD8+ T cells with unpulsed or hTERT461-469 peptide-loaded (1 μM) K562-A24, K562, HLA-A*24:02+, or HLA-A*24:02− allogeneic B-LCLs at the indicated effector:target (E/T) ratios. (C) Effect of HLA class I and class II blockade on the cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against the cognate peptide-pulsed (1 μM) K562-A24 was determined by 51Cr-release assays at an E/T ratio of 5:1. (D) hTERT-siTCR–transduced CD8+ T cells were tested in 51Cr release assays against K562 (negative control) and K562-A24 cells pulsed with the indicated concentrations of hTERT461-469 peptide at an E/T ratio of 5:1. Error bars represent SDs. (E) Representative flow cytometry plots showing staining of K3-1 with the HLA-A*24:02/hTERT461-469 tetramer (upper) and the irrelevant HLA-A*24:02/HIV-1 Env584-592 tetramer (negative control; bottom). (F) IFN-γ production by hTERT-siTCR–transduced CD8+ T cells was measured by using a format similar to that described for panel D. The parental K3-1 CTL clone was tested in parallel.
hTERT-siTCR–transduced CD8+ T cells display epitope-specific responsiveness. (A) Representative flow cytometry plots showing staining of hTERT-siTCR–transduced CD8+ T cells with HLA-A*24:02/hTERT461-469 tetramer. HLA-A*24:02/HIV tetramer was used as a negative control. (B) 51Cr-release assays were conducted by using hTERT-siTCR–transduced CD8+ T cells with unpulsed or hTERT461-469 peptide-loaded (1 μM) K562-A24, K562, HLA-A*24:02+, or HLA-A*24:02− allogeneic B-LCLs at the indicated effector:target (E/T) ratios. (C) Effect of HLA class I and class II blockade on the cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against the cognate peptide-pulsed (1 μM) K562-A24 was determined by 51Cr-release assays at an E/T ratio of 5:1. (D) hTERT-siTCR–transduced CD8+ T cells were tested in 51Cr release assays against K562 (negative control) and K562-A24 cells pulsed with the indicated concentrations of hTERT461-469 peptide at an E/T ratio of 5:1. Error bars represent SDs. (E) Representative flow cytometry plots showing staining of K3-1 with the HLA-A*24:02/hTERT461-469 tetramer (upper) and the irrelevant HLA-A*24:02/HIV-1 Env584-592 tetramer (negative control; bottom). (F) IFN-γ production by hTERT-siTCR–transduced CD8+ T cells was measured by using a format similar to that described for panel D. The parental K3-1 CTL clone was tested in parallel.
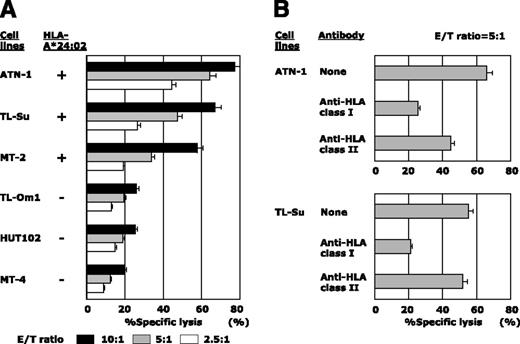
hTERT-siTCR/CD8 dose-dependently killed the HLA-A*24:02+ ATL/HTLV-I–infected cell lines ATN-1, TL-Su, and MT-2, but not the HLA-A*24:02− TL-Om1, HUT102, and MT-4 (Figure 4A). Additionally, the tumoricidal effect mediated by hTERT-siTCR/CD8 was abrogated by anti-HLA class I mAb, but not by anti-HLA-DR mAb (Figure 4B). Furthermore, time-lapse imaging directly demonstrated this tumoricidal activity of hTERT-siTCR/CD8 against HLA-A*24:02+ ATN-1, but not that against HLA-A*24:02− HUT102 or K562 (negative control) (supplemental Fig 1-(1)). We then examined the tumoricidal activity against freshly isolated ATL tumor cells and found that these transfectants also dose-dependently killed HLA-A*24:02+, but not -A*24:02− freshly isolated ATL tumor cells (Figure 5A).
Cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against ATL/HTLV-I–infected cell lines. (A) Cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against HLA-A*24:02+ or HLA-A*24:02− ATL/HTLV-I–infected cell lines was tested in 51Cr-release assays at the indicated E/T ratios. All tested ATL/HTLV-I–infected cell lines overexpressed hTERT mRNA and protein, as shown in Figure 1. (B) Effect of HLA class I and class II blockade on the cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against ATN-1 and TL-Su was tested in 51Cr-release assays at an E/T ratio of 5:1.
Cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against ATL/HTLV-I–infected cell lines. (A) Cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against HLA-A*24:02+ or HLA-A*24:02− ATL/HTLV-I–infected cell lines was tested in 51Cr-release assays at the indicated E/T ratios. All tested ATL/HTLV-I–infected cell lines overexpressed hTERT mRNA and protein, as shown in Figure 1. (B) Effect of HLA class I and class II blockade on the cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells against ATN-1 and TL-Su was tested in 51Cr-release assays at an E/T ratio of 5:1.
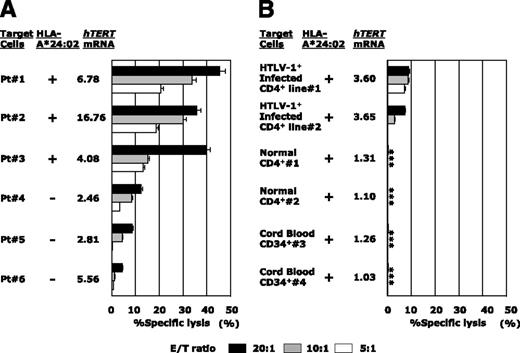
hTERT-siTCR–transduced CD8+ T cells kill freshly isolated ATL cells and newly HTLV-I–infected CD4+ T cells, but not normal cells, in vitro. (A) Freshly isolated HLA-A*24:02+ (n = 3) or HLA-A*24:02− (n = 3) ATL tumor cells overexpressing hTERT mRNA were used as targets in 51Cr-release assays with hTERT-siTCR–transduced CD8+ T cells at the indicated E/T ratios. (B) The same hTERT-siTCR–transduced CD8+ T cells used in panel A at the same E/T ratios were tested in 51Cr-release assays against newly generated HLA-A*24:02+ HTLV-I–infected CD4+ T cells (n = 2) representing HTLV-I carrier CD4+ T cells, original HLA-A*24:02+ normal CD4+ T cells (n = 2) representing the normal counterpart ATL tumor cells (corresponding number indicating cells from the identical donor), and HLA-A*24:02+ normal CB-CD34+ cells (n = 2) encompassing steady-state normal hematopoietic progenitor cells. Listed levels of expression of hTERT mRNA are those relative to the mean levels of expression across 6 PBMC samples from healthy donors determined by qRT-PCR and calculated by using the comparative threshold cycle method. Error bars represent SDs (* indicates less than detectable).
hTERT-siTCR–transduced CD8+ T cells kill freshly isolated ATL cells and newly HTLV-I–infected CD4+ T cells, but not normal cells, in vitro. (A) Freshly isolated HLA-A*24:02+ (n = 3) or HLA-A*24:02− (n = 3) ATL tumor cells overexpressing hTERT mRNA were used as targets in 51Cr-release assays with hTERT-siTCR–transduced CD8+ T cells at the indicated E/T ratios. (B) The same hTERT-siTCR–transduced CD8+ T cells used in panel A at the same E/T ratios were tested in 51Cr-release assays against newly generated HLA-A*24:02+ HTLV-I–infected CD4+ T cells (n = 2) representing HTLV-I carrier CD4+ T cells, original HLA-A*24:02+ normal CD4+ T cells (n = 2) representing the normal counterpart ATL tumor cells (corresponding number indicating cells from the identical donor), and HLA-A*24:02+ normal CB-CD34+ cells (n = 2) encompassing steady-state normal hematopoietic progenitor cells. Listed levels of expression of hTERT mRNA are those relative to the mean levels of expression across 6 PBMC samples from healthy donors determined by qRT-PCR and calculated by using the comparative threshold cycle method. Error bars represent SDs (* indicates less than detectable).
Conversely, as shown in Figure 5B, neither HLA-A*24:02+ normal CD4+ T cells (the normal counterpart of ATL tumor cells) nor HLA-A*24:02+ normal CB-CD34+ cells as normal hematopoietic progenitor cells were killed. In the same experiment, newly established IL-2–dependent HTLV-I–infected CD4+ T cells (Patient #1 and Patient #2), but not the corresponding original normal/HTLV-I− CD4+ T cells (Patient #1 and Patient #2), became to some extent sensitive to the same transfectants as the level of hTERT mRNA expression increased (Figure 5B). This observation confirmed that not only ATL tumor cells, but also HTLV-I–infected cells from which ATL tumor cells were derived could be killed by these hTERT-specific effector cells.
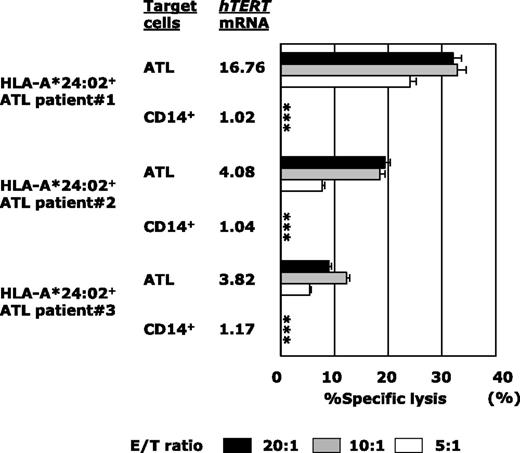
Next, because the majority of ATL patients were of an advanced age and were therefore ineligible for allo-HSCT, we examined the tumoricidal activity against autologous ATL tumor cells mediated by gene-modified PB-CD8+ T cells from the patient (Figure 6). Although PB-CD8+ T cells from heavily pretreated ATL patients were sometimes difficult to subject to TCR gene modification and ex vivo expansion, hTERT-siTCR/CD8 cells generated from HLA-A*24:02+ patients (n = 3) were able to substantially lyse autologous ATL tumor cells in proportion to the corresponding level of hTERT mRNA expression. Autologous CD14+ PB monocytes were used as a negative control because they lacked expression of hTERT mRNA. These results demonstrated that hTERT-siTCR/CD8 cells were able to exert tumoricidal activity against ATL tumor cells through recognition of the hTERT461-469 epitope/HLA-A*24:02 complex, which is naturally presented on the surface of ATL tumor cells.
hTERT-siTCR–transduced CD8+ T cells kill freshly isolated autologous ATL tumor cells on the basis of hTERT expression levels. Cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells obtained from HLA-A*24:02+ ATL patients (n = 3) against autologous freshly isolated ATL tumor cells and autologous peripheral CD14+ cells (negative control) was tested in 51Cr-release assays at the indicated E/T ratios. hTERT mRNA in each patient’s ATL tumor cells is listed using a format similar to that used in Figure 5. Error bars represent SDs (* indicates less than detectable).
hTERT-siTCR–transduced CD8+ T cells kill freshly isolated autologous ATL tumor cells on the basis of hTERT expression levels. Cytotoxic activity of hTERT-siTCR–transduced CD8+ T cells obtained from HLA-A*24:02+ ATL patients (n = 3) against autologous freshly isolated ATL tumor cells and autologous peripheral CD14+ cells (negative control) was tested in 51Cr-release assays at the indicated E/T ratios. hTERT mRNA in each patient’s ATL tumor cells is listed using a format similar to that used in Figure 5. Error bars represent SDs (* indicates less than detectable).
hTERT-siTCR–transduced CD8+ T cells display in vivo anti-ATL reactivity
In vivo anti-ATL reactivity mediated by hTERT-siTCR/CD8 cells was assessed by using a xenografted mouse model and bioluminescence assay. Serial bioluminescence assay images were simultaneously acquired.
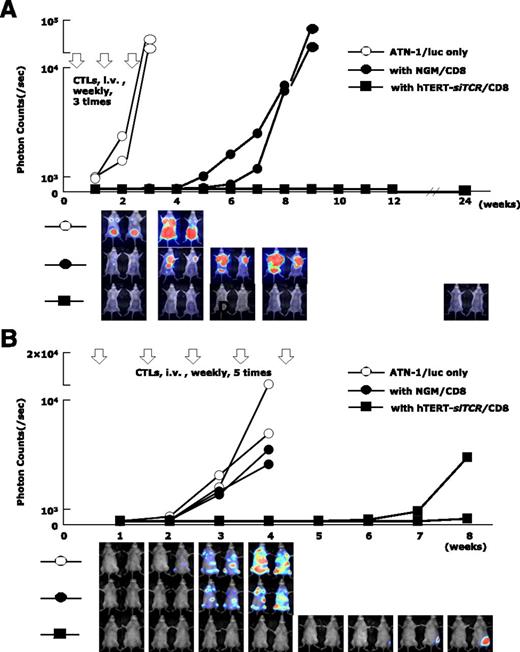
In the Winn assay (Figure 7A), tumor cell growth in NOG mice treated with hTERT-siTCR/CD8 (n = 2) was completely inhibited for longer than 6 months. In contrast, when compared with nontreated NOG mice (n = 2) in which the inoculated ATL tumor mass rapidly enlarged, activated NGM/CD8 (n = 2) did suppress ATL tumor growth to some degree, but eventually huge tumor masses developed within 2 months. In a therapeutic adoptive transfer model (Figure 7B), the tumor cell growth in mice treated with hTERT-siTCR/CD8 (n = 2) was obviously suppressed within the 8-week observation period, in contrast to that in mice treated with NGM/CD8 (n = 2) and that in control mice (n = 2).
Anti-ATL reactivity of hTERT-siTCR–transduced CD8+ T cells in vivo. (A) Winn assay. NOG mice were coinjected with a luciferase-transduced HLA-A*24:02+ ATL cell line (ATN-1/luc) (5 × 105) and either 2.5 × 106 hTERT-siTCR–transduced (hTERT-siTCR/CD8) or NGM/CD8+ T cells (n = 2 per group). Subsequently, 3 weekly infusions of the respective CD8+ T-cell populations (2.5 × 106 cells per infusion) were administered intravenously (i.v.). Tumor growth was monitored every 7 days by using bioluminescence assay. Nontreated ATN-1/luc cells were similarly inoculated into NOG mice (n = 2) as a control. Although NGM/CD8 activated using OKT-3 and rhIL-2 suppressed tumor growth to some extent, hTERT-siTCR/CD8 durably suppressed tumor growth for longer than 6 months. (B) Therapeutic adaptive transfer model. NOG mice were intravenously inoculated with 5 × 105 ATN-1/luc cells. Four days later, intravenous administration of either 5 × 106 hTERT-siTCR/CD8 or NGM/CD8 (n = 2 per group) was started once a week for a total of 5 infusions. NOG mice given only ATN-1/luc cells (n = 2) were used as a control. In comparison with NGM/CD8, therapeutically infused hTERT-siTCR/CD8 also obviously suppressed the tumor cell growth within the 8-week observation period. Serial images of the bioluminescence assay demonstrate tumor growth in each group.
Anti-ATL reactivity of hTERT-siTCR–transduced CD8+ T cells in vivo. (A) Winn assay. NOG mice were coinjected with a luciferase-transduced HLA-A*24:02+ ATL cell line (ATN-1/luc) (5 × 105) and either 2.5 × 106 hTERT-siTCR–transduced (hTERT-siTCR/CD8) or NGM/CD8+ T cells (n = 2 per group). Subsequently, 3 weekly infusions of the respective CD8+ T-cell populations (2.5 × 106 cells per infusion) were administered intravenously (i.v.). Tumor growth was monitored every 7 days by using bioluminescence assay. Nontreated ATN-1/luc cells were similarly inoculated into NOG mice (n = 2) as a control. Although NGM/CD8 activated using OKT-3 and rhIL-2 suppressed tumor growth to some extent, hTERT-siTCR/CD8 durably suppressed tumor growth for longer than 6 months. (B) Therapeutic adaptive transfer model. NOG mice were intravenously inoculated with 5 × 105 ATN-1/luc cells. Four days later, intravenous administration of either 5 × 106 hTERT-siTCR/CD8 or NGM/CD8 (n = 2 per group) was started once a week for a total of 5 infusions. NOG mice given only ATN-1/luc cells (n = 2) were used as a control. In comparison with NGM/CD8, therapeutically infused hTERT-siTCR/CD8 also obviously suppressed the tumor cell growth within the 8-week observation period. Serial images of the bioluminescence assay demonstrate tumor growth in each group.
Discussion
Although ATL still has a poor prognosis, the clinical presence of the graft-versus-ATL in patients treated successfully by allo-HSCT has encouraged the search for a novel cellular immune-mediated treatment of ATL. Unlike EBV-related malignancy,6 the feasibility of HTLV-I–associated Tax7 and HBZ8 proteins as therapeutic targets of anti-ATL immunotherapy still remains controversial. Therefore, in this study, we explored the feasibility of a novel therapeutic target other than one associated with HTLV-I. Consequently, we demonstrated for the first time that hTERT was a promising therapeutic target for anti-ATL adoptive immunotherapy. Freshly isolated ATL tumor cells produced hTERT mRNA abundantly, and HLA-A*24:02-restricted and hTERT461-469-specific CTL precursors were detected in the periphery of HLA-A*24:02+ ATL patients. These findings suggested that naturally processed and presented hTERT461-469/HLA-A*24:02 complex on the surface of ATL tumor cells was sufficiently immunogenic to be recognized by the target-specific CTLs in HLA-A*24:02+ ATL patients. Additionally, hTERT mRNA expression in newly generated HTLV-I–infected CD4+ T cells was upregulated, and these cells became sensitive to gene-modified hTERT-specific CTLs (Figure 5B). The involvement of Tax12 and HBZ38 in upregulation of the hTERT gene in HTLV-I–infected immortalized CD4+ T cells and ATL tumor cells has been reported previously. Initially, it might seem more realistic to develop an hTERT461-469 peptide vaccine for treatment of HLA-A*24:02+ ATL patients. However, because we were concerned that CTL induction of hTERT peptide vaccine might have a tendency to be impeded by the regulatory T-cell function of ATL tumor cells,21 we focused on developing a redirected T-cell–based adoptive immunotherapy targeting hTERT to allow administration of a number of hTERT-specific CTLs directly.
To this end, we cloned the full-length rearranged TCR-α/β genes from K3-1, the HLA-A*24:02-restricted and hTERT461-469-specific CTL clone.19 With codon optimization of the constant regions, we inserted them into our new souped-up second-generation 2A peptide-based siTCR vector to accomplish an increased expression level of the introduced TCR, carrying small interfering RNAs for the endogenous TCR-α/β genes in the redirected T cells (hTERT-siTCR vector).26,27,34 The siTCR vector system makes it possible to simultaneously accomplish profound suppression of endogenous TCR genes and markedly increase the cell-surface expression of the introduced TCR, resulting in upregulated antitumor reactivity,34 thus leading to inhibition of mispaired TCR formation between the endogenous and introduced TCR-α and -β chains, and lowering the potential risk of lethal graft-versus-host disease.39 We found that both allogeneic and autologous gene-modified CD8+ T cells using the hTERT-siTCR vector successfully killed ATL tumor cells both in vitro and in vivo (Figures 4-7), but not normal cells, including steady-state hematopoietic progenitor cells (Figure 5B). The introduced cytocidal activity against ATL tumor cells mediated by these gene-modified CTLs was actually accomplished through recognition of the HLA-A*24:02/hTERT461-469 complex on the surface of ATL tumor cells (Figures 3 and 4).
Clinical studies of anticancer immunotherapy targeting hTERT have not demonstrated any significant adverse events so far.14-17,20 However, for clinical application, because a number of activated gene-modified hTERT-specific CTLs would be administered at once, it would again be necessary to be mindful of on-target adverse events against normal tissues that constitutively express the hTERT gene.10,40 Notably, any impairment of hematopoiesis would be the major concern. In this study, both allogeneic and autologous gene-modified effector CD8+ T cells expressing hTERT-specific TCR from adult peripheral lymphocytes, and CB lymphocytes did not kill CB-CD34+ cells representing steady-state hematopoietic progenitors (Figure 5B). By using cytokine-driven myeloid differentiation with CB-CD34+ cells, gene-modified CTLs targeting hTERT showed a slight cytocidal effect against differentiated and highly proliferating subsets of CD34+CD33+ and CD34−CD33+ cells but spared CD34+CD33dim cells (supplemental Fig 2A). Additionally, contrary to resting CD4+ cells and CD19+ cells, highly mitotic polyhydroxic acid–stimulated CD4+ cells and CD19+ EBV LCLs became sensitive to effector CTLs because of increased expression of hTERT mRNA, the latter being more salient (Figure 5B and supplemental Fig 2B). Taken together, our findings suggest that gene-modified hTERT-specific CTLs will spare steady-state hematopoietic progenitor cells. However, to ensure safety, it would be better to avoid the active recovery phase of bone marrow after chemotherapy, notably under granulocyte colony-stimulating factor support, and also the acute infectious period in which immune-cell components are stimulated.
Another likely problem in clinical practice is that heavily pretreated peripheral lymphocytes from ATL patients might fail to proliferate. Proliferative activity of therapeutically infused gene-modified T cells in vivo is an important prerequisite for a successful outcome.41 In this connection, although the control of treatment-related graft-versus-host disease still remains unsolved, use of CB lymphocytes has been investigated.42 In this study, gene-modified CB-CD8+ T cells from 2 donors successfully killed ATL tumor cells but spared autologous steady-state CB-CD34+ cells (supplemental Figure 1-(2)). Compelling lack of suitable allo-HSCT donors for patients of advance age with ATL will encourage the application of CB transplantation using reduced-intensity preconditioning in the near future. Genetic redirection of CB lymphocytes using tumor antigen–specific TCR gene transfer will also play a considerable role.
Conversely, because hTERT is overexpressed in various kinds of cancer,10 this approach may have widespread potential clinical application. Furthermore, the clinical availability of a new defucosylated anti-CCR4 mAb for treatment of ATL43 can be reasonably anticipated to diminish regulatory T cells, the key player in the immunosuppressive microenvironment in patients with cancer,44 because CCR4 is also expressed on regulatory T cells.45 Therefore, hTERT-targeting immunotherapy after preconditioning with this anti-CCR4 mAb may become a realistically promising treatment option not only for ATL, but also for other malignancies.
In summary, using a newly established hTERT-siTCR vector, we have demonstrated the feasibility of anti-ATL redirected T-cell–based adoptive immunotherapy targeting hTERT, notably for patients who are ineligible for allo-HSCT. Further studies will be needed to investigate the clinical safety and utility of this novel therapy.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful for the skilled technical assistance of Dr Kenji Kameda, Ehime University, and Dr Hirofumi Inoue, Department of Biochemistry and Molecular Genetics, Ehime University Graduate School of Medicine. Thanks are also extended to Dr Yoshiki Akatsuka, Department of Hematology, Fujita Health University, for supplying the K562-A*24:02 cell line, Dr Midori Okumura and Dr Tomihiro Katayama, Department of Obstetrics and Gynecology, Ehime University Graduate School of Medicine, for supplying cord blood samples, and Dr Hiroo Saji, HLA Laboratory, Japan, for HLA typing.
This work was supported in part by grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (H.F., T.A., and M.Y.), a Grant-in-Aid for Cancer Research from the Ministry of Health, Labor and Welfare (M.Y.), and a grant from the Japan Leukemia Research Fund (2011) (H.F.).
Authorship
Contribution: Y.M. performed the research and wrote the paper; H.F. designed and performed the research, wrote and edited the paper and provided financial support; H.A., F.O., and T.O. performed the research and discussed the experimental results; T.A. interpreted the experimental results and provided financial support; T.I., S.O., J.M., K.K., and H.S. provided materials and discussed the experimental results; and M.Y. discussed and interpreted the experimental results and provided financial support.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Hiroshi Fujiwara, Department of Bioregulatory Medicine, Ehime University Graduate School of Medicine, Toon, Ehime 791-0295, Japan; e-mail: yunarief@m.ehime-u.ac.jp.

![Figure 1. Abundant expression of hTERT in ATL tumor cells. (A) Expression of hTERT mRNA in ATL/HTLV-I infected cell lines (▪), freshly isolated ATL tumor cells from patients (▲), normal PBMCs (○), and CB-CD34+ cells (△) were examined by qRT-PCR. The level of hTERT mRNA expression in the K562 leukemia cell line (●) was used as an internal control. The expression level of hTERT mRNA in each sample was calculated relative to that of PBMCs. hTERT mRNA expression relative to normal PBMCs was 21.3 ± 17.9 for the ATL/HTLV-I–infected cell line, 7.48 ± 6.89 for freshly isolated ATL tumor cells, and 1.10 ± 0.12 for CB-CD34+ (mean ± standard deviation [SD]). The ATL/HTLV-I–infected cell line and freshly isolated ATL tumor cells expressed hTERT mRNA abundantly and significantly (*P < .01). (B) Expression of hTERT protein in ATL cell lines and normal PBMCs was confirmed by western blotting.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/121/24/10.1182_blood-2012-11-465971/4/m_4894f1.jpeg?Expires=1769162414&Signature=2mjmF5Z77Yqyodo8KPGXgTdXua-K2NeGw7n3xR~SHHWfhu~5I9JKFy59Vh1lvfsFcapUfLFvSKEk2kQDpuqffYHNMJRCSLYWmXU834WrvyTJ~eORVQFj6HQjNKzNQMiSELjmDlNuqLnxS13UEdXr2bbG~gLrkxtVWqoFn7SicE--M3dcFnZuvC8jOcHbB4CC7m5LOSJYLqgEEDzaLKAJR1gfcAQrUmehqWjeu~jOaW-7Ti0EQsAENaXAV2j7X3CrwG5mZv4-tcbl0H1jaX0L7xO8KfA8iwjDxDQHJtMiy1NY~vbQv6YSQ-FaGXPEmaNowD19jRvIZ2oNCroGuH-HtQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal