Key Points
The CUL4 ubiquitin ligases target HOXB4 for ubiquitination and degradation.
A degradation-resistant HOXB4 variant markedly enhanced ex vivo expansion of adult hematopoietic stem cells.
Direct transduction of the homeobox (HOX) protein HOXB4 promotes the proliferation of hematopoietic stem cells (HSCs) without induction of leukemogenesis, but requires frequent administration to overcome its short protein half-life (∼1 hour). We demonstrate here that HOXB4 protein levels are post-translationally regulated by the CUL4 ubiquitin ligase, and define the degradation signal sequence (degron) of HOXB4 required for CUL4-mediated destruction. Additional HOX paralogs share the conserved degron in the homeodomain and are also subject to CUL4-mediated degradation, indicating that CUL4 likely controls the stability of all HOX proteins. Moreover, we engineered a degradation-resistant HOXB4 that conferred a growth advantage over wild-type HOXB4 in myeloid progenitor cells. Direct transduction of recombinant degradation-resistant HOXB4 protein to human adult HSCs significantly enhanced their maintenance in a more primitive state both in vitro and in transplanted NOD/SCID/IL2R-γnull mice compared with transduction with wild-type HOXB4 protein. Our studies demonstrate the feasibility of engineering a stable HOXB4 variant to overcome a major technical hurdle in the ex vivo expansion of adult HSCs and early progenitors for human therapeutic use.
Introduction
Hematopoietic stem cells (HSCs) are pluripotent, asymetrically self-renewing cells that give rise to all mature blood cells through successive rounds of differentiation. The signals that govern the self-renewal process have been intensively pursued, as it may be possible to promote HSC expansion by transiently enforcing proliferation pathways or blocking differentiation cues, which would be highly desirable as a means to augment the number of HSCs for transplantation. Currently, HSC transplantation is used to treat hematologic diseases, such as leukemia and bone marrow failure. HSCs are derived from 3 sources: umbilical cord blood (UCB), adult bone marrow, and peripheral blood after treatment with recombinant human granulocyte colony-stimulating factor (G-CSF), a cytokine that mobilizes HSCs from the bone marrow and allows their harvest through peripheral blood apheresis. Ex vivo expansion of HSCs from these sources would improve transplantation outcomes and help meet the demand for stem cell transplants by permitting the use of samples of limited quantity (eg, cord blood) or with low total numbers of HSCs (eg, poor HSC mobilizers).
Ex vivo expansion of HSCs and progenitor cells has been the subject of considerable investigation with the use of combinations of hematopoietic growth factors,1 synthetic small molecular weight compounds identified by high-throughput screening,2 and the use of supportive bone marrow stroma.3,4 In human studies, adult HSCs can undergo repeated rounds of asymmetric self-renewal with maintenance of the stem cell pool but with little or no expansion. In contrast, fetal and neonatal stem cells can be maintained in culture for 2 to 3 months with absolute increases in the number of HSCs. Ex vivo expansions of sevenfold to ninefold in SCID-repopulating HSCs have been reported over 12 to 14 days in a number of studies.1,3,5
Homeobox (HOX) genes encode transcription factors that regulate patterning during embryonic development and hematopoiesis, both prenatally and post-natally. In early development, HOX gene expression is both temporally and spatially regulated, as reflected by the sequential order of transcription with respect to their 3′ to 5′ chromosomal position. However, the spatiotemporal regulation of HOX gene expression is not observed in hematopoiesis, but instead assumes a complex, overlapping expression pattern that is not lineage-specific. Northern blotting analysis revealed that most HOX genes, excluding HOXD, are highly expressed in HSCs and progenitor cells, and are generally down-regulated as cells terminally differentiate into mature, lineage-specific blood cells.6 To date, the mechanisms underlying transcriptional regulation of HOX genes during hematopoiesis remain largely unknown.
The identification of HOX genes that are highly expressed in CD34+ HSCs and early progenitors led to several overexpression studies that revealed the effects of specific HOX genes on HSC proliferation. In particular, sustained HOXB4 expression by retroviral transduction promoted the selective expansion of murine HSCs in cell culture and after bone marrow transplantation.7 Moreover, the enhanced proliferation of HOXB4-transduced HSCs was not leukemogenic in transplanted mice,8,9 indicating that enforced HOXB4 expression did not alter HSC differentiation. In marked contrast, the ectopic expression of other HOX genes highly expressed in HSCs, such as HOXA9 and HOXB7, did result in myeloid leukemia.10,11 Comparison of the effects of HOXB4 overexpression in different species further revealed species-specific variations in the magnitude of HOXB4-induced HSC proliferation, with more modest effects observed in cells from humans and baboons than mice and dogs.12 Thus, the effect of ectopic HOXB4 expression in promoting human HSC proliferation is likely more equivocal than the robust outcome obtained in murine HSCs.
Follow-up studies revealed the development of myeloid leukemia in large mammals 2 years post-transplantation with HSCs expressing retrovirally transduced HOXB4,12 highlighting the risks of such genetic manipulation. Although the molecular basis underlying the observed leukemogenesis appears to be HOXB4-dependent, integration of a transgene at chromosomal hotspots may also lead to the activation of adjacent oncogenes, and is believed to contribute to gene therapy-associated malignancy.13,-15 Therefore, a transient, nonviral delivery method to augment HOXB4 levels in HSCs is necessary for potential human therapeutic applications. To that end, direct protein transduction has been established as an alternative method of introducing HOXB4 into HSCs.16,17 Comparable levels of ex vivo murine HSC proliferation were achieved with the protein transduction of recombinant human HOXB4 as well as with retroviral integration,17 but the short half-life of HOXB4 necessitated frequent replenishing of the recombinant protein, which is impractical for large-scale ex vivo expansion of HSCs. Studies using human embryonic stem cells18 or bone marrow cells19,20 also demonstrate the effects of recombinant HOXB4 protein for therapeutic use. Stabilization of the HOXB4 protein through post-translational manipulation would represent a major advance in providing a renewable source of HSCs for human therapeutic use.
We previously studied the post-translational regulation of homeodomain (HD)-containing proteins by the CUL4A ubiquitin ligase, and we determined that CUL4A targets HOXA9, a transcription factor that promotes HSC proliferation, for ubiquitination and proteosome-mediated degradation.21 In addition, the AML-associated Nup98-HOXA9 fusion protein is resistant to CUL4A-mediated ubiquitination, and the resulting protein stabilization may contribute to its potent leukemogenic activity.22 In this study, we showed that representative members of multiple HOX paralogous groups, including HOXB4, are subjected to ubiquitination and subsequent degradation by CUL4A. The degradation signal in HOXB4 was identified as the LEXE motif in helix I of the HD, which is conserved in all 39 HOX HD proteins. Mutation of this motif resulted in the profound stabilization of HOXB4 protein levels. Retroviral transduction of 32D cells, a murine myeloid progenitor cell line, with degradation-resistant HOXB4 mutant, dramatically enhanced their proliferation potential compared with wild-type HOXB4. Finally, the addition of recombinant degradation-resistant HOXB4 protein to adult HSCs resulted in higher numbers of colony-forming cells in vitro and remarkable efficiency in the maintenance of primitive CD34+ cells in the engrafted bone marrow of NOD/SCID/IL2R-γnull (NSG) mice.
Methods
Cell cultures, plasmids, and protein extracts
Steady-state levels of epitope-tagged HOX proteins in response to increasing levels of MYC-CUL4A were determined after transfection of 293T cells with 1 μg of the indicated HOX plasmids (lanes 1 to 4) and 1, 3, or 9 μg MYC-CUL4A (lane 2 to 4). All DNA amounts were normalized with vector DNA, and protein levels were detected using standard western blot techniques with anti-MYC (Roche, Indianapolis, IN), anti-HA (Covance, Princeton, NJ), and anti-β-actin (Santa Cruz, Dallas, TX) antibodies.
To measure the effect of CUL4A silencing on HOXB4 half-life, HeLa cells were transiently transfected with 6 μg MYC-HOXB4 and 6 μg short hairpin (sh) control or 6 μg shCUL4A. Pulse-chase of transfected HeLa cells was performed as previously described.23 The half-lives of 35S-labeled-MYC-HOXB4 and HOXB4 degron mutants were determined by immunoprecipitation and immunoblotting with anti-MYC antibody, and were quantitated via phosphorimaging.
To determine the effect of CUL4A inhibition on the ubiquitination of MYC-HOXB4, 293T cells were transiently transfected with 3 μg MYC-HOXB4 (lanes 1 to 5), 3 μg HIS-ubiquitin (lanes 2 to 5), and 1, 3, or 9 μg V5-tagged dominant negative CUL4A (DN-CUL4A, lanes 3 to 5).23 Transfected cells were treated with 25 μM MG132 for 4 hours prior to harvesting. Cell lysates were precipitated with Ni2+-NTA agarose beads (Qiagen, Valencia, CA), and immunoblotted with antibodies against MYC, V5 (Invitrogen, Grand Island, NY), and α-tubulin (Sigma, St Louis, MO).
To determine the effect of the CUL4 ubiquitin ligases on endogenous HOXB4 protein levels in hematopoietic progenitor cells, bone marrow cells from wild-type and null Cul4a and Cul4b mice were isolated, and progenitor cells were enriched using the EasySep Mouse Hematopoietic Progenitor Cell Enrichment Kit (Stemcell Technologies, Vancouver, BC, Canada). Genotyping was performed as previously described.24 HOXB4 protein levels were analyzed by western blot analysis using standard techniques with an anti-HOXB4 antibody (Cell Signaling, Danvers, MA).
Mutations of the HOXB4 LEXE motif were generated using the Quikchange Multi Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). The eGFP-HOXB4 HD (wild-type and mutant) fusion constructs were generated by polymerase chain reaction amplification of wild-type and mutant HOXB4 HD and ligation to pEGFP-C2 (BD Bioscience, San Jose, CA). Steady-state levels of eGFP-HOXB4 HD in response to increasing levels of MYC-CUL4A expression were determined after transfection of 293T cells with 1 μg eGFP-HOXB4 WT HD (lanes 1 to 4) or 1 μg eGFP-HOXB4 M3 HD (lanes 5 to 8), together with 1 μg (lanes 2 and 6), 3 μg (lanes 3 and 7), or 9 μg (lanes 4 and 8) MYC-CUL4A. All DNA amounts were normalized with vector DNA, and protein levels were detected using standard western blot techniques.
Wild-type and mutant HA-HOXB4 were retrovirally transduced into 32D cells as previously described,21 and were sorted by GFP expression. The MIGRI retroviral vector alone (lacking a transgene) was also transduced into 32D cells as a negative control. Ectopic expression of wild-type and mutant HA-HOXB4 were confirmed by standard western blot techniques. The stable lines were cultured in Iscove's modified Dulbecco's medium (IMDM, Mediatech, Manassas, VA) supplemented with 20% Knockout Serum Replacement (Invitrogen), 1% penicillin/streptomycin (Mediatech, Manassas, VA), and 2 ng/mL murine interleukin (IL)-3 (Miltenyi Biotech, Auburn, CA). Cells were passaged every 3 days and counted daily. HOXB4 half-life was measured after addition of 50 μM cycloheximide (final concentration). Samples were harvested at the indicated times, and HA-HOXB4 levels were determined by western blot analysis.
In vitro HSC assays
To express recombinant protein, wild-type and mutant HOXB4 were subcloned into the pTAT-HA expression vector (generous gift from Dr Steven Dowdy). Recombinant protein was purified as previously described,18 and was desalted using PD-10 columns (GE Healthcare Life Sciences, Pittsburgh, PA). G-CSF–mobilized CD34+ cells were grown in serum-free QBSF medium (Quality Biological, Gaithersburg, MD) supplemented with 100 ng/mL thrombopoietin (Miltenyi Biotech), c-Kit ligand (Amgen, Thousand Oaks, CA) and Flt3 ligand (Amgen), and 1 μg/mL recombinant protein every other day. Colony-forming cell assays were performed with G-CSF-mobilized CD34+ cells cultured in triplicate (500 cells/plate). Each plate contained 1 mL of semisolid medium with 1.2% methylcellulose (Corning Chemicals, Corning, NY), 20% Serum Replacement, 80 μM 2-mercaptoethanol, 2 mM l-glutamine, 1% penicillin/streptomycin, 0.125 mM hemin (Sigma), 20 ng/mL G-CSF (Amgen), c-Kit ligand and IL-3 (Amgen), and 6 U/mL erythropoietin (Amgen) and 1 μg/mL recombinant proteins. Colonies were scored after 2 weeks of incubation at 37°C.
Limiting dilution cobblestone area-forming colony (CAFC) assays were performed in 96-well format with the indicated numbers of G-CSF–mobilized CD34+ cells added to MS-5 stromal cells concurrently with transplantation into NSG mice. Cultures were changed weekly with the replacement of one-half of the culture volume with fresh media (MEMα [Memorial Sloan-Kettering Cancer Center] supplemented with 12.5% horse serum [HyClone, Rockford, IL], 12.5% fetal calf serum, 80 μM b2-mercaptoethanol, 2 mM l-glutamine, 1% penicillin/streptomycin and 1 μM hydrocortisone [Sigma]). After 5 weeks, CAFCs were scored using an inverted-phase microscope as phase-dark hematopoietic areas of at least 5 cells beneath the MS-5 stromal layer.
Transplantation experiments
NSG mice bred in the MSKCC animal facility were irradiated at 250 cGy (cesium source). Each irradiated mouse (5 mice/group) was transplanted via tail vein injection with 3 × 105 CD34+ G-CSF–mobilized adult HSCs (AllCells, LLC, Emeryville, CA) treated with 2 rounds of purified recombinant wild-type or degradation-resistant HOXB4 protein (1 μg/mL) added immediately after and 24 hours after overnight cytokine stimulation (100 ng/mL thrombopoietin, c-Kit ligand, and Flt3 ligand). Mice were sacrificed at 12 weeks post-transplantation, and bone marrow was harvested for fluorescence-activated cell sorting analysis using monoclonal antibodies against human CD19, CD33, CD34, and CD45 (Miltenyi Biotech). All protocols were Institutional Review Board (IRB) and Institutional Animal Care and Use Committee (IACUC) approved.
Results
CUL4A mediates ubiquitination of HOXB4
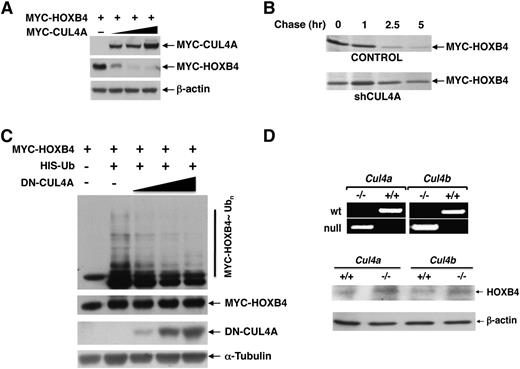
We previously determined that HOXA9 is specifically targeted for ubiquitination and degradation by CUL4A,21 and observed that members of the HOX4 family also interacted with CUL4A by yeast 2-hydrid assay (Zhang and Zhou, unpublished). Due to its well-established ability to promote HSC self-renewal, we first evaluated whether HOXB4 is subject to CUL4A-mediated degradation. The steady-state levels of MYC-tagged HOXB4 decreased corresponding to increasing levels of MYC-CUL4A (Figure 1A). Conversely, knockdown of CUL4A by sh RNA prolonged the half-life of HOXB4 (Figure 1B and supplemental Figure 1). After the addition of MG132, we observed that HOXB4 is polyubiquitinated, and enforced expression of dominant-negative CUL4A (previously described in Chen et al23 ) dramatically reduced ubiquitination of HOXB4 in a dose-dependent manner (Figure 1C).
CUL4A targets HOXB4 for ubiquitin-dependent degradation. (A) Steady-state levels of MYC-HOXB4 were determined in response to increasing levels of cotransfected MYC-CUL4A in 293T cells by immunoblotting with the anti-MYC and β-actin (loading control) antibodies. (B) Pulse-chase analysis of HOXB4 half-life was performed following shCUL4A knockdown. (C) In vivo ubiquitination of HOXB4 in response to increasing levels of dominant-negative CUL4A was detected in 293T cells after tranfection with the indicated plasmids, treatment with MG132, precipitation with Ni2+-NTA agarose beads under denaturing conditions, and immunoblotting with anti-MYC antibody. (D) To determine the physiological effect of CUL4A and CUL4B on endogenous HOXB4 protein levels, bone marrow stem and progenitor cells from Cul4a and Cul4b knockout mice and their wild-type littermates were purified and analyzed by immunoblotting with anti-HOXB4. The upper panel shows the genotyping of Cul4a−/− and Cul4b−/Y mice by polymerase chain reaction.
CUL4A targets HOXB4 for ubiquitin-dependent degradation. (A) Steady-state levels of MYC-HOXB4 were determined in response to increasing levels of cotransfected MYC-CUL4A in 293T cells by immunoblotting with the anti-MYC and β-actin (loading control) antibodies. (B) Pulse-chase analysis of HOXB4 half-life was performed following shCUL4A knockdown. (C) In vivo ubiquitination of HOXB4 in response to increasing levels of dominant-negative CUL4A was detected in 293T cells after tranfection with the indicated plasmids, treatment with MG132, precipitation with Ni2+-NTA agarose beads under denaturing conditions, and immunoblotting with anti-MYC antibody. (D) To determine the physiological effect of CUL4A and CUL4B on endogenous HOXB4 protein levels, bone marrow stem and progenitor cells from Cul4a and Cul4b knockout mice and their wild-type littermates were purified and analyzed by immunoblotting with anti-HOXB4. The upper panel shows the genotyping of Cul4a−/− and Cul4b−/Y mice by polymerase chain reaction.
CUL4A and CUL4B are 82% identical, and target similar substrates for ubiquitination and degradation.24,25 To determine the role of CUL4A and CUL4B on HOXB4 stability in hematopoietic progenitor cells, we compared the endogenous HOXB4 protein levels in hematopoietic stem and progenitor cells derived from Cul4a- and Cul4b-null mice vs their wild-type littermates, and observed that the deletion of either Cul4a or Cul4b resulted in the increased accumulation of HOXB4 protein levels (Figure 1D). Taken together, these results indicate a role for both CUL4A and CUL4B as regulators of HOXB4 stability. Our studies hereafter used CUL4A as the representative CUL4 E3 ligase to investigate the biochemical mechanisms and functional implications of this E3 family in orchestrating the ubiquitination and degradation of HOXB4.
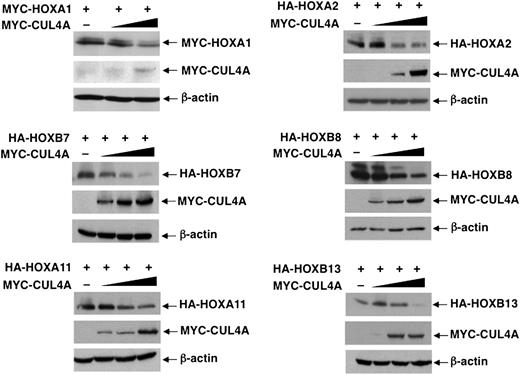
With the evidence that CUL4A targets both HOXA9 and HOXB4 for ubiquitination, next we sought to determine whether other HOX proteins in the class I HD family are subject to CUL4A-mediated degradation. Indeed, the steady-state levels of representative HOX paralogous group proteins, including HOXA1, A2, B7, B8, A11, and A13, also decreased corresponding to increasing levels of MYC-CUL4A (Figure 2), whereas the HOX cofactor Meis1 was not subjected to CUL4A-mediated degradation (supplemental Figure 2). Thus, the post-translational regulation of HOX protein stability by the CUL4A ubiquitin ligase may be mediated through a conserved mechanism, such as recognition of a common HOX protein domain.
CUL4A targets multiple HOX proteins for degradation. Steady-state levels of indicated HOX proteins were determined in response to increasing levels of cotransfected CUL4A, as detected by immunoblotting with the appropriate antibodies. β-actin levels were determined by immunoblotting as loading controls.
CUL4A targets multiple HOX proteins for degradation. Steady-state levels of indicated HOX proteins were determined in response to increasing levels of cotransfected CUL4A, as detected by immunoblotting with the appropriate antibodies. β-actin levels were determined by immunoblotting as loading controls.
CUL4A-specific HOXB4 degron
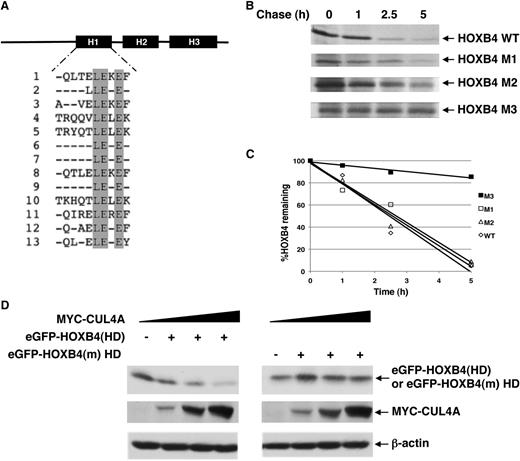
Our previous studies on HOXA9 indicated that the HD that is required for DNA-binding activity of HOX proteins is also necessary for CUL4A-mediated HOXA9 turnover.21 A sequence alignment of the highly conserved HD among all HOX proteins revealed a conserved LEXE motif in helix I that is not involved in DNA binding (Figure 3A).26 Given our observation that several HOX paralogous groups are post-translationally regulated by CUL4A, we examined whether the conserved LEXE motif may comprise the CUL4A-dependent degron or the substrate sequence that directs its recognition by a specific ubiquitin ligase for degradation. Although mutation of the first 2 residues of the LEXE motif prolonged the HOXB4 protein half-life compared with wild-type protein, the triple mutant, HOXB4(M3), dramatically extended HOXB4 protein stability (Figure 3B-C). Accordingly, we designated the triple mutant as degradation-resistant HOXB4 or HOXB4(m).
HOXB4 HD contains the CUL4A-dependent degron. (A) A sequence alignment of the HDs of all HOX paralogous groups revealed a conserved LEXE motif. (B-C) Half-lives of HOXB4 LEXE mutants. Pulse-chase analysis of HOXB4 LEXE mutants (M1: L175D; M2: L175D, E176K; M3: L175D, E176K, E178K) was performed after transfection of the MYC-tagged proteins into HeLa cells, 35S-Met labeling, and immunoprecipitation with the anti-MYC antibody. The percentages of HOXB4 and the LEXE mutant proteins remaining at each time point were normalized relative to time 0 of each protein. The percentages of HOXB4 or LEXE mutant proteins remaining at each time point are indicated. (D) Determination of the steady-state levels of GFP-HOXB4 HD fusion protein in response to increasing MYC-CUL4A levels by immunoblotting with antibodies against GFP or MYC epitope tags, with β-actin as loading control.
HOXB4 HD contains the CUL4A-dependent degron. (A) A sequence alignment of the HDs of all HOX paralogous groups revealed a conserved LEXE motif. (B-C) Half-lives of HOXB4 LEXE mutants. Pulse-chase analysis of HOXB4 LEXE mutants (M1: L175D; M2: L175D, E176K; M3: L175D, E176K, E178K) was performed after transfection of the MYC-tagged proteins into HeLa cells, 35S-Met labeling, and immunoprecipitation with the anti-MYC antibody. The percentages of HOXB4 and the LEXE mutant proteins remaining at each time point were normalized relative to time 0 of each protein. The percentages of HOXB4 or LEXE mutant proteins remaining at each time point are indicated. (D) Determination of the steady-state levels of GFP-HOXB4 HD fusion protein in response to increasing MYC-CUL4A levels by immunoblotting with antibodies against GFP or MYC epitope tags, with β-actin as loading control.
To determine if the LEXE motif in the context of its native tertiary structure is required to target proteins for CUL4A-mediated degradation, wild-type or mutant HOXB4 HD was fused to eGFP, a long-lived protein that is not a native CUL4A substrate. The fusion of eGFP to the wild-type HOXB4 HD conferred sensitivity to CUL4A-mediated degradation, whereas the eGFP-HOXB4 mutant HD protein remained resistant to increasing levels of CUL4A (Figure 3D). We conclude from these studies that the LEXE motif within helix I of the HOXB4 HD constitutes a transferable signal that is both necessary and sufficient to mediate CUL4A-dependent degradation.
HOXB4(m) enhances 32D proliferation
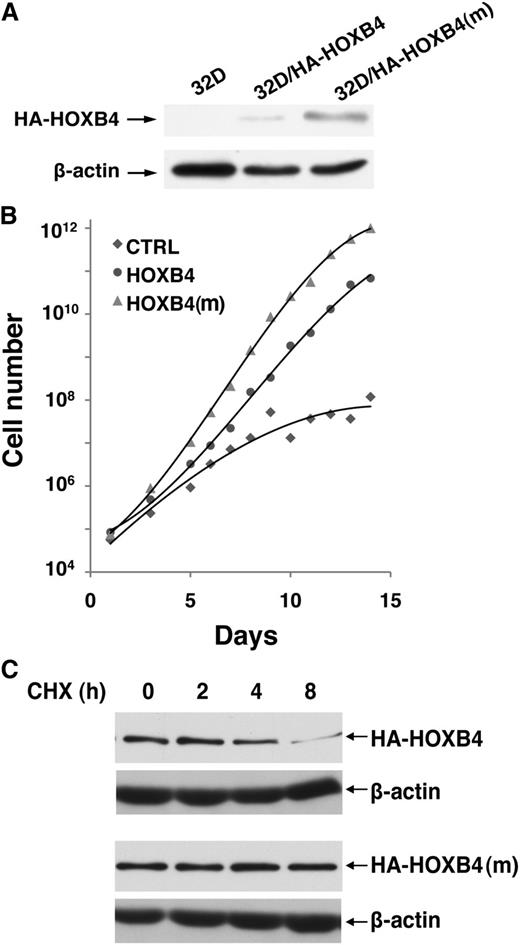
To determine whether HOXB4 degradation by CUL4A impacts the proliferation potential of hematopoietic cells, we retrovirally transduced HOXB4 or HOXB4(m) into the 32D murine myeloid progenitor cell line and observed that steady-state levels of HOXB4(m) were higher than wild-type HOXB4, whereas the mRNA levels were comparable (Figure 4A and supplemental Figure 3). Next, we compared their growth in serum-free medium and found that ectopic expression of either wild-type or degradation-resistant HOXB4 resulted in a distinct growth advantage over control cells. Remarkably, 32D cells expressing HOXB4(m) showed a further 10-fold increase in cell number compared with wild-type HOXB4-transduced cells after 15 days in culture, representing a 10 000-fold increase compared with 32D cells transduced with vector alone (Figure 4B). Correspondingly, the half-life of HOXB4(m) was substantially longer than wild-type HOXB4 (Figure 4C), indicating that the enhanced protein stability of HOXB4(m) contributes to the growth advantage of 32D myeloid progenitors.
Degradation-resistant HOXB4 confers a proliferative advantage to the 32D myeloid cell line. (A) Expression levels of HA-HOXB4 (wild-type or mutant) in 32D stable lines, as determined by immunoblotting with the anti-HA antibody. (B) Cumulative expansion of 32D stable lines in serum-free medium. (C) Cycloheximide chase analysis for the half-lives of HA-HOXB4 (wild-type or mutant) in 32D stable lines. HA-HOXB4 or HA-HOXB4(m) levels were determined by immunoblotting at the indicated time points. β-actin levels were determined as the loading control.
Degradation-resistant HOXB4 confers a proliferative advantage to the 32D myeloid cell line. (A) Expression levels of HA-HOXB4 (wild-type or mutant) in 32D stable lines, as determined by immunoblotting with the anti-HA antibody. (B) Cumulative expansion of 32D stable lines in serum-free medium. (C) Cycloheximide chase analysis for the half-lives of HA-HOXB4 (wild-type or mutant) in 32D stable lines. HA-HOXB4 or HA-HOXB4(m) levels were determined by immunoblotting at the indicated time points. β-actin levels were determined as the loading control.
HOXB4(m) maintains HSC potential
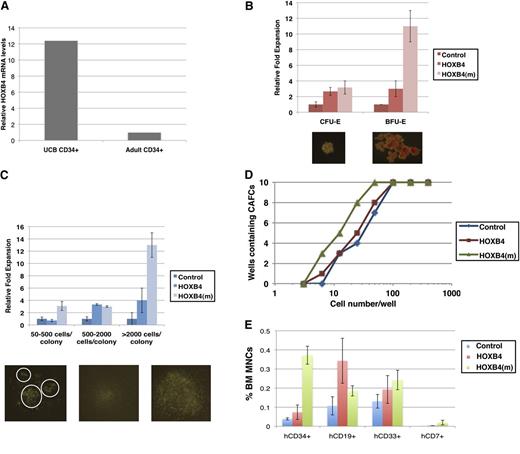
HSCs derived from peripheral blood are more readily available and abundant than those derived from UCB for stem cell transplantation. To determine the effect of HOXB4 on postnatal HSC expansion, we first compared the expression of HOXB4 in CD34+ cells from cord blood vs G-CSF–mobilized adult peripheral blood. As determined by quantitative RT-qPCR in Figure 5A, HOXB4 mRNA levels are 12-fold lower in G-CSF–mobilized adult CD34+ cells than UCB CD34+ cells, suggesting that HOXB4 may be a limiting factor in the ex vivo expansion of G-CSF–mobilized adult HSCs, and that ectopic introduction of HOXB4 protein into G-CSF–mobilized adult CD34+ cells may promote more dramatic HSC expansion than UCB CD34+ cells.
Direct transduction of recombinant degradation-resistant HOXB4 protein maintains G-CSF–mobilized CD34+ cells in a more primitive state than wild-type HOXB4. (A) Comparison of HOXB4 mRNA expression levels in CD34+ UCB cells vs CD34+ G-CSF–mobilized cells. (B-C) Comparison of granulocyte-monocyte colony-forming cells (CFU-GM), erythroid burst-forming cells (BFU-E), and erythroid colony-forming cells (CFU-E) after addition of wild-type or degradation-resistant recombinant HOXB4 protein to CD34+ G-CSF–mobilized cells. (D) Limiting dilution CAFC assays of G-CSF–mobilized CD34+ cells after addition of wild-type or degradation-resistant HOXB4 protein. (E) Multi-lineage engraftment of G-CSF–mobilized CD34+ cells treated with wild-type or degradation-resistant recombinant HOXB4 protein in NSG mice (n = 8 per group) 12 weeks post-primary transplantation. The experiment was independently repeated 3 times.
Direct transduction of recombinant degradation-resistant HOXB4 protein maintains G-CSF–mobilized CD34+ cells in a more primitive state than wild-type HOXB4. (A) Comparison of HOXB4 mRNA expression levels in CD34+ UCB cells vs CD34+ G-CSF–mobilized cells. (B-C) Comparison of granulocyte-monocyte colony-forming cells (CFU-GM), erythroid burst-forming cells (BFU-E), and erythroid colony-forming cells (CFU-E) after addition of wild-type or degradation-resistant recombinant HOXB4 protein to CD34+ G-CSF–mobilized cells. (D) Limiting dilution CAFC assays of G-CSF–mobilized CD34+ cells after addition of wild-type or degradation-resistant HOXB4 protein. (E) Multi-lineage engraftment of G-CSF–mobilized CD34+ cells treated with wild-type or degradation-resistant recombinant HOXB4 protein in NSG mice (n = 8 per group) 12 weeks post-primary transplantation. The experiment was independently repeated 3 times.
Our goal of engineering the degradation-resistant HOXB4(m) protein is to overcome the labile nature of HOXB4 (t1/2 ∼1 hour) used in protein-based ex vivo expansion of HSC and progenitor cells.17 To this end, HOXB4 and HOXB4(m) were tagged with the cell-penetrating HIV TAT protein transduction domain that allows efficient translocation across cellular membranes (reviewed in Schwarze et al 27 and references therein). It is also noteworthy that the HD of HOXB4 possesses membrane-penetrating activity.16 TAT-HA-HOXB4 and TAT-HA-HOXB4(m) were expressed as hexahistidine-tagged fusions in Escherichia coli, affinity-purified, and applied to G-CSF–mobilized adult CD34+ cells for evaluation in clonogenic assays. Cell entry and perinuclear localization of recombinant HOXB4 and HOXB4(m) protein were observed in G-CSF–mobilized CD34+ cells (supplemental Figure 4), consistent with the typical punctate distribution of transduced TAT-fusion proteins.28 Diffuse nuclear localization was also observed, especially for TAT-HA-HOXB4(m) (supplemental Figure 4). The transcriptional activity of putative HOXB4 target genes was similar after the addition of wild-type or degradation-resistant HOXB4 protein (supplemental Figure 5). As shown in Figure 5B-C, the total number of both myeloid and erythroid colony-forming cells increased >10-fold with the addition of HOXB4(m) compared with wild-type HOXB4 protein. Moreover, myeloid granulocyte-monocyte colony-forming cells and erythroid burst-forming cells grown in the presence of HOXB4(m) were significantly larger and more frequent (Figure 5B-C). In contrast, UCB CD34+ cells responded similarly after the addition of recombinant wild-type or degradation-resistant HOXB4 protein (supplemental Figure 6), further highlighting the effect of HOXB4(m) in G-CSF–mobilized adult HSCs. We also performed limiting dilution CAFC assays to quantify the number of HSCs in G-CSF–mobilized adult CD34+ cells that were treated with either wild-type or degradation-resistant HOXB4 recombinant protein. Untreated cells served as the baseline control. Addition of degradation-resistant HOXB4(m) protein doubled the number of HSCs measured by limiting dilution compared with both control and G-CSF–mobilized CD34+ cells treated with wild-type HOXB4 protein (Figure 5D), indicating that the prolonged half-life of HOXB4 recombinant protein increased the efficiency in maintaining and expanding G-CSF–mobilized HSCs ex vivo.
To determine the effect of wild-type vs degradation-resistant HOXB4 protein on the engraftment of G-CSF–mobilized adult HSCs and progenitors, CD34+ cells were treated with recombinant protein, transplanted into sublethally irradiated NSG mice, and analyzed 10 weeks post-transplantation. HOXB4(m) protein significantly enhanced the number of human CD34+ cells in the bone marrow of primary transplanted mice compared with both control and wild-type HOXB4 protein-treated mice, and multi-lineage engraftment was also observed (Figure 5E). Strikingly, a fivefold increase in the percentage of primitive CD34+ HSCs and progenitors was detected in the engrafted bone marrow of HOXB4(m)-transduced NSG mice. Thus, the degradation-resistant HOXB4(m) protein enhances proliferation of G-CSF–mobilized HSCs in vitro and maintains the primitive state of HSCs more effectively than wild-type HOXB4 in vivo.
Discussion
Previous studies have demonstrated the ability of HOXB4 to expand HSCs, but a practical method of delivering HOXB4 had yet to be devised that would ensure safety and pharmaceutical compatibility. Although the ectopic expression of HOXB4 after retroviral transduction was not reported to induce leukemia in mice, more recent studies in large mammals found a causal relationship between the development of leukemia and their transplantation with HSCs constitutively expressing HOXB4.12 Direct transduction of wild-type HOXB4 protein recapitulated the expansion of HSCs similar to enforced expression of the HOXB4 gene, but required frequent replenishing of the recombinant protein due to the short protein half-life.17 Subsequent studies have indicated a longer protein half-life is possible with the truncation of the HOXB4 N-terminus, but the exact mechanism that accounts for this effect is unknown.18 In this study, we have determined the biochemical mechanism for the post-translational regulation of HOXB4 that can be exploited to advance the field of ex vivo HSC expansion. We previously demonstrated that HOXA9 was targeted for CUL4A-mediated degradation, and found that the degradation signal resided in the HD.21 Here, we determined that CUL4A is responsible for the proteasome-mediated degradation of multiple HOX proteins, and identified a conserved HD motif as a significant determinant for CUL4A-directed activity. Fusion of the HOXB4 HD to a non-native CUL4A substrate was sufficient to target the protein for CUL4A-mediated degradation, indicating that the HOXB4 HD contains a bona fide CUL4A degron. Future studies may exploit this finding to direct the degradation of proteins in a CUL4A-dependent manner.
Constitutive Cul4a- and Cul4b-knockout mice exhibited no defects in hematopoiesis.24,25 Here, we showed that the deletion of either of the CUL4 family members led to increased accumulation of HOXB4, indicating a redundant role for CUL4A and CUL4B in vivo. Future experiments are required to address the regulatory mechanisms that trigger CUL4-mediated degradation of HOX proteins, which may have significant effects on the proliferation and/or differentiation of hematopoietic cells. Quantitative reverse-transcription polymerase chain reaction revealed higher levels of HOXB4 mRNA in CD34+ UCB cells than adult CD34+ cells, which may account for the modest effect of exogenous HOXB4 protein on UCB CD34+ cells compared with G-CSF–mobilized adult CD34+ progenitor cells in colony-forming cell assays. Moreover, degradation-resistant HOXB4 enhanced the proliferation of the murine myeloid progenitor 32D cell line and the generation of more primitive myeloid and erythroid colonies by G-CSF CD34+ progenitor cells compared with wild-type HOXB4, further indicating that the effect of degradation-resistant HOXB4 may be augmented in hematopoietic progenitor cells with limiting levels of HOXB4.
More than 50 000 HSC transplants are performed each year worldwide,29 of which the vast majority use adult HSCs mobilized into the peripheral blood after treatment with G-CSF.30 HSCs from UCB or adult G-CSF–mobilized bone marrow/peripheral blood are not equivalent in their transplantation outcome. Cord blood is limited in the number of CD34+ stem/progenitor cells that can be obtained (1 to 2 × 106), an amount sufficient for the engraftment of children or small adults. For larger adults, the use of 2 cord blood units has proven effective but requires a double match and considerable increased cost. Transplantation with HSCs derived from a sibling or unrelated matched bone marrow or G-CSF–mobilized peripheral blood donors requires a minimum of 2 × 106 CD34+ cells/kg. In the case of poor mobilizers, combinations of AMD3100 and G-CSF can be used to increase CD34+ yield and/or multiple leukaphereses may be undertaken. Another disadvantage of using G-CSF–mobilized CD34+ cells for transplantation is the time required to identify a suitable match and to schedule the harvest. In light of the unstable nature of recombinant wild-type HOXB4 protein that impedes the development of protein-based ex vivo expansion of adult HSCs and progenitors, our studies represent an important advance in elucidating a post-translational regulatory mechanism of HOXB4 protein stability that may be exploited to maintain the proliferative and repopulative potential of HSCs and early progenitors. Up to one-third of autologous transplant patients require additional rounds of G-CSF treatment to mobilize sufficient numbers of HSCs for transplant. We demonstrated that degradation-resistant HOXB4 protein showed a stronger effect in expanding G-CSF–mobilized HSCs, which may be used to eliminate the need for repeat HSC mobilization procedures. Future studies will seek to fully reconstitute the HOXB4 ubiquitination reaction in vitro, which may offer mechanistic insight into the specific inhibition of HOXB4 degradation to promote the expansion of hematopoietic stem and progenitor cells.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Lorraine Gudas (Weill Cornell Medical College), Steve Dowdy, and Mark Kamps (University of California, San Diego) for plasmids, Lisa Prevedel for technical assistance, and Jeffrey Hannah for critical reading of the manuscript.
This work is supported by grants from the National Institutes of Health (CA118085 and CA098210), and National Institute of Allergy and Infectious Diseases, the Leukemia and Lymphoma Society Scholar Award, the Irma T. Hirschl Career Scientist Award (P.Z.), National Institute of Allergy and Infectious Diseases (HHSN272200900059C) (M.A.S.M.), the Italian Association for Cancer Research (IG-9204) (G.M.), the Roberta C. Rudin Leukemia Research Fund, and the Gar Reichman Fund of the Cancer Research Institute.
Authorship
Contribution: J.L. and J.-H.S. designed and performed research, and analyzed and interpreted the data; J.Z., L.L., Y.Z., and J.Y.E. performed part of the assays; P.Z. and M.A.S.M. designed and directed the research and provided funding; G.M. contributed to data analysis and interpretation; J.L. and P.Z. wrote the manuscript; and G.M. and M.A.S.M. edited the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pengbo Zhou, Department of Pathology and Laboratory Medicine, Weill Cornell Medical College, 1300 York Ave, New York, NY 10065; e-mail: pez2001@med.cornell.edu.