Key Points
Demonstrates efficient reprogramming of iPS cells from CD34+ stem cells enriched from a small volume of peripheral blood.
Abstract
A variety of somatic cells can be reprogrammed to induced pluripotent stem cells (iPSCs), but CD34+ hematopoietic stem cells (HSCs) present in nonmobilized peripheral blood (PB) would be a convenient target. We report a method for deriving iPSC from PB HSCs using immunobead purification and 2- to 4-day culture to enrich CD34+ HSCs to 80% ± 9%, followed by reprogramming with loxP-flanked polycistronic (human Oct4, Klf4, Sox2, and c-Myc) STEMCCA-loxP lentivector, or with Sendai vectors. Colonies arising with STEMCCA-loxP were invariably TRA-1-60+, yielding 5.3 ± 2.8 iPSC colonies per 20 mL PB (n = 17), where most colonies had single-copy STEMCCA-loxP easily excised by transient Cre expression. Colonies arising with Sendai were variably reprogrammed (10%-80% TRA-1-60+), with variable yield (6 to >500 TRA-1-60+ iPSC colonies per 10 mL blood; n = 6). Resultant iPSC clones expressed pluripotent cell markers and generated teratomas. Genomic methylation patterns of STEMCCA-loxP–reprogrammed clones closely matched embryonic stem cells. Furthermore, we showed that iPSCs are derived from the nonmobilized CD34+ HSCs enriched from PB rather than from any lymphocyte or monocyte contaminants because they lack somatic rearrangements typical of T or B lymphocytes and because purified CD14+ monocytes do not yield iPSC colonies under these reprogramming conditions.
Introduction
Induced pluripotent stem cells (iPSCs) are a valuable research tool and may in the future be an unlimited source of autologous cells differentiated from iPSCs for treatments. To date, iPSCs have been made from numerous somatic cell types including dermal fibroblasts,1-3 lymphocytes,4,5 mesenchymal stem cells,6 and mobilized CD34+ cells.7 Although coding mutations have been identified in fibroblast-derived iPSCs,8 CD34+ hematopoietic stem cells (HSCs) may maintain greater genomic stability than terminally differentiated somatic cells,9 making them attractive targets for iPSC reprogramming. Furthermore, iPSCs derived from HSCs may be predisposed toward redifferentiation to HSCs via “epigenetic memory”,10,11 making such HSC-derived iPSCs particularly suitable for development of research models and treatments for hematopoietic diseases.
Human HSCs are multipotent adult stem cells associated with the cell surface marker CD34. Previous reports have used granulocyte CSF (G-CSF) mobilized CD34+ cells7 or peripheral blood mononuclear cells (PBMCs) for iPSC reprogramming.12 Hematopoietic stem/progenitor stem CD34+ cells are present in low numbers in circulating peripheral blood13 due to steady state mobilization from bone marrow. Here we demonstrate iPSC derivation from CD34+ cells enriched from small volumes of peripheral blood by venipuncture from healthy donors and patients without G-CSF mobilization, a relatively noninvasive and easily accessible cell source.
Methods
Human subjects and animal use approvals
CD34+-enriched HSCs used in this study were obtained from 6 healthy volunteer donors and 22 patients with primary immune deficiencies under study in our National Institutes of Health (NIH) program. We obtained written informed consent from all subjects following the Declaration of Helsinki under the auspices of National Institute of Allergy and Infectious Diseases (NIAID) Institutional Review Board–approved protocol 05-I-0213 and also from 3 donors under protocol 94-I-0073. The donors not treated with any mobilizing agent had peripheral blood obtained in EDTA or heparin tubes by venipuncture, and for 3 of these patients PBMCs were cryopreserved before thaw for use in the study. The donors participating in protocol 94-I-0073 underwent 5 days of G-CSF 16 µg/kg mobilization and apheresis collection of an HSC-enriched leukocyte fraction.
Immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJNSG (NSG) mice (The Jackson Laboratory) were used for teratoma assays of iPSC pluripotency, as approved by the NIAID Institutional Animal Care and Use Committee under animal use protocol LHD 3E.
CD34+ cell purification and culture
PBMCs were purified from nonmobilized peripheral blood by Ficoll separation (Lymphocyte Separation Medium; MP Biomedicals). Similar results were obtained when CD34+ cells were isolated from PBMCs using either CD34 antibody-conjugated Dynal (Life Technologies) or MACS (Miltenyi Biotec) magnetic beads according to each manufacturer’s protocol. The apheresis product from patients mobilized with G-CSF was processed using the CliniMAX CD34+ cell separation system (Miltenyi) in the NIH Department of Transfusion Medicine, Cell Processing Section. Nonmobilized CD34+ cells were expanded for 2 to 4 days to obtain approximately 50 000 cells in HSC media consisting of X-VIVO 10 media (Biowhittaker) with 1% human serum albumin (Talecris Biotherapeutics); 50 ng/mL each of human stem cell factor, Flt3-ligand, and thrombopoietin; and 5 or 50 ng/mL of human interleukin-3 (PeproTech). Characterization of cell lineages obtained after purification and HSC media culture was assessed by flow cytometry with fluorescence-conjugated antibodies for CD3, CD14, CD19, and CD34 expression (BD Biosciences).
STEMCCA-loxP virus production
STEMCCA-loxP lentiviral vector (polycistronic encoding human Oct4, Klf4, Sox2, and c-Myc) was produced through transient transfection of 293T cells with hSTEMCCA-loxP14 plasmid (gift from Dr Gustavo Mostoslavsky) and gag-pol, rev-tat, and VSV-G packaging plasmids.15 Viral vector was harvested daily for 3 days, filtered (0.22 μm; Millipore), and concentrated by centrifugation at 18 600g for 4 hours at 4°C, then reconstituted in X-VIVO 10 media. Although we produced our own lentivector, ready-to-use STEMCCA-loxP is available in kit format (Millipore).
qPCR for lentivirus titer analysis
RNA was isolated from lentivirus vector preparations using High Pure Viral RNA kit (Roche). Complementary DNA (cDNA) was synthesized from viral RNA using High Capacity RNA-to-cDNA kit (Life Technologies). To measure lentivirus titer, quantitative polymerase chain reaction (qPCR) was performed using SYBR green dye (Maxima SYBR Green/ROX qPCR Master Mix; Fermentas) in an Applied Biosystems 7500 Real-Time PCR System. Lentivirus-specific primers between U5 and gag/pol gene were used (FWD Primer #7: 5′-ACTCGGCTTGCTGAAGCGCGCACGGCAAGAGG-3′ and BWD Primer #8: 5′-CCATCGCGATCTAATTCTCCCCCGCTTAATAC-3′). HIV-lentiviral vector plasmid DNA was diluted in log-scale (ranging from 102 to 108 copies) for standard curve. STEMCCA-loxP virus titer was adjusted relative to the known titer of a GFP lentivirus measured by both qPCR and flow cytometry in transduced K562 cells (ATCC).
Overview of STEMCCA-loxP reprogramming
The timeframe for iPSC generation and characterization is CD34+ cell transduction and iPSC generation (3-4 weeks), iPSC expansion (3 weeks), Cre-excision of STEMCCA-loxP (1 week), postexcision iPSC expansion (3 weeks), in vitro embryoid body (EB) 3-germ layer differentiation (2-3 weeks), and in vivo teratoma 3-germ layer differentiation (6-12 weeks).
STEMCCA-loxP transduction and iPSC derivation
CD34+ cells were transduced once overnight with STEMCCA-loxP lentivirus at a multiplicity of infection (MOI) of 2 viruses per cell on RetroNectin (Lonza)-coated plates with protamine sulfate (1-5 µg/mL) by spinoculation at 2500 rpm for 30 minutes at 32°C and then returned to 37°C culture. The next morning, cells were exchanged into fresh HSC media and 2 days later were transferred to mitotically inactive primary CF-1 mouse embryonic fibroblasts (MEFs; Millipore) in HSC media. The next day, media was replaced with standard iPSC media (3 mL/well for 6-well plates) consisting of KnockOut Dulbecco’s modified Eagle medium (DMEM)/F12 medium (Life Technologies), 20% KnockOut Serum Replacement (Life Technologies), 1% L-glutamine (Quality Biological), 1% penicillin-streptomycin (Life Technologies), 1% nonessential amino acids (Life Technologies), 0.1 mM β-mercaptoethanol (Sigma-Aldrich), and 10 ng/mL human basic fibroblast growth factor (PeproTech). iPSC media was changed every 2 days until iPSC colonies were mechanically plucked (typically 18-21 days after transduction) based on colony morphology and positive staining for TRA-1-60 pluripotency marker (described below). Colonies were transferred into individual wells of 96-well plates containing fresh MEFs in iPSC media containing 10 μM ROCK inhibitor (Y-27632; EMD Biochemicals or Sigma-Aldrich) for reduced apoptosis on the day of passaging. The next day, media was replaced with iPSC media lacking ROCK inhibitor, which was changed daily. iPSC colonies were expanded for additional pluripotency characterization, with passages of 1:2 or 1:4 into larger wells, using culture conditions described below.
CytoTune Sendai reprogramming vectors transduction and iPSC derivation
PB-derived CD34+ cells were cotransduced by spinoculation with the 4 CytoTune Sendai viruses separately encoding hOct4, hKlf4, hSox2, and hc-Myc (Life Technologies) at an MOI of 6 for each virus (a higher MOI than manufacturer recommended) and then cultured overnight, followed by media change. Then 2 to 3 days after transduction, cells were plated in iPSC media onto MEFs. After 7 additional days, cells were passaged 1:6 onto fresh MEFs; additional MEFs were added only as needed if the MEF layer showed any signs of degradation before the colonies that arose were ready to be plucked. The number of TRA-1-60+ colonies that arose were scored, and some were plucked for characterization.
iPSC culture
iPSCs were cultured and characterized using standard techniques.6,16 Specifically, iPSCs were cultured at 37°C with 5% CO2 on MEFs in standard iPSC media (described above) changed daily during routine culture. iPSCs were passaged mechanically or enzymatically using 1 mg/mL collagenase (Life Technologies) and were plated onto fresh MEFs in iPSC media containing 10 μM ROCK inhibitor on the day of passaging.
iPSC staining for pluripotency markers
For live cell stain for TRA-1-60, 1:200 dilutions of primary antibody (mouse anti-human TRA-1-60; BD Biosciences) and secondary antibody (Alexa Fluor 555–conjugated goat anti-mouse immunoglobulin M; Life Technologies) in iPSC media were added directly to the culture, and incubated for 1 hour at 37°C. Wells were then washed twice with PBS, fresh iPSC media was added, and colonies were visualized by fluorescence microscopy.
For fixed cell staining, cells were fixed with 4% paraformaldehyde (Wako Chemicals) for 30 minutes, washed with PBS, and incubated with Perm/Wash buffer (BD Biosciences). Fixed cells were stained for TRA-1-60 (as above), washed twice, stained with Hoechst dye (1:10 000 dilution for 1 minute; Life Technologies), and analyzed by fluorescence microscopy. Separate wells of fixed cells were stained using an alkaline phosphatase (AP) detection kit (Millipore) according to the manufacturer’s protocol.
STEMCCA-loxP vector copy number analysis in iPSCs
STEMCCA-loxP vector integration in iPSCs was quantified using genomic DNA purified from 1 to 2 million cells using QIAamp DNA Mini Kit (QIAGEN) and diluted to 20 ng/mL. TaqMan real-time qPCR was performed using the following primers: LentiGAG-Probe 5′-6FAM-ACAGCCTTCTGATGTTTCTAACAGGCCAGG-NFQ-MGB-3′, LentiGAG-F 5′-GGAGCTAGAACGATTCGCAGTTA-3′, LentiGAG-R 5′-GGTTGTAGCTGTCCCAGTATTTGTC-3′, with TaqMan RNaseP assay (Life Technologies) as the genomic reference standard.
LAM-PCR
To identify STEMCCA-loxP vector integration sites, linear amplification mediated (LAM) PCR17 insertion site analysis was performed using the following primers: Linker cassette forward: 5′-(Phos)-GACCCGGGAGATCTGAATTCAGTGGCACAGCAGTTAGG-3′, Linker cassette reverse: 5′-(Phos)-AATTCCTAACTGCTGTGCCACTGAATTCAGATC-3′, LC I: 5′-GACCCGGGAGATCTGAATTC-3′, LC II: 5′-GATCTGAATTCAGTGGCACAG-3′ and LV-LTR1: 5′-(biotin)-GAGCTCTCTGGCTAACTAGG-3′, LV-LTR2: 5′-AGCTTGCCTTGAGTGCTTCA-3′, LV-LTR3: 5′-AGTAGTGTGTGCCCGTCTGT-3′. LAM-PCR products were purified using QIAGEN gel purification kit and directly sequenced with LC2 and LV-LTR3 primers.
EB differentiation
Confluent iPSCs in 3 wells of a 6-well plate were detached from MEFs with dispase (1 mg/mL for 20-30 minutes at 37°C; Life Technologies). Cells were washed 3 times and cultured as nonadherent EBs in 3 mL EB medium per well of 6-well ultra–low-attachment plates (Corning) for 7 to 10 days. EBs were then harvested and broken into clumps and then replated onto 3 gelatin-coated wells (24-well plate) for 2 to 4 days of adherent growth. EB medium consisted of KnockOut DMEM/F12, 20% fetal bovine serum (HyClone), 1% L-glutamine, 1% penicillin-streptomycin, and 1% nonessential amino acids. After EB differentiation, cells were fixed and permeabilized in wells (as described above) and stained at room temperature in Perm/Wash buffer containing unconjugated primary antibody to α-fetoprotein (1:100 dilution; Abcam) or smooth muscle actin (1:100 dilution; Millipore) for 1 hour, or with Alexa Fluor 555–conjugated class III β-tubulin (1:10 dilution; BD Biosciences) for 2 hours, followed by 2 washes. Unconjugated primary antibodies were detected with secondary Alexa Fluor 555–conjugated anti-mouse immunoglobulin G or immunoglobulin G2a antibodies (1:200 dilution for 1 hour; Life Technologies). Cells were counterstained with Hoechst dye (1:10 000 dilution for 1 minute) and visualized by fluorescence microscopy.
Teratoma formation
Pluripotency of iPSCs was tested in vivo by teratoma assay (either before and/or after Cre excision for clones produced with STEMCCA-loxP). For iPSC harvest, cells were incubated with 1 mg/mL collagenase for 5 minutes, wells were washed twice with PBS, and cells were detached using a cell scraper. Then 6 to 8 wells (6-well plates) of iPSCs were harvested for each mouse to be injected. Cells were resuspended in precooled (4°C) 100 µL KnockOut DMEM/F12 + 100 µL hESC-qualified Matrigel (BD Biosciences) and kept on ice until intramuscular injection into the hindlimb quadricep of NSG mice using a 23G to 25G needle. Teratomas typically formed within 2 to 3 months after injection and were excised, fixed in 4% paraformaldehyde, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E; American HistoLabs).
Puro-T2A-Cre-GFP construct
For convenience, we used a lentivector transfer plasmid (pCL20i4r-EF1a-Puro-T2A-Cre-GFP) that we constructed containing puromycin resistance and Cre-GFP fusion protein (from Addgene plasmid 13776)18 genes coexpressed as a single T2A-linked transcript from a short elongation factor-1α promoter. Our initial plan had been to use this lentivector transfer plasmid to produce replication-incompetent lentivector for Cre-mediated excision of STEMCCA-loxP, but to avoid any possibility of genomic insertion of this vector we simply used the vector plasmid DNA to transfect the targeted iPSC clones and followed the transient fluorescence of the Cre-GFP fusion protein as a marker of the success of this approach.
STEMCCA-loxP excision
To prevent preferential transfection of MEFs during STEMCCA-loxP excision, iPSCs were cultured under feeder-free conditions on Matrigel-coated 12-well plates in mTeSR1 (Stem Cell Technologies) or NutriStem XF/FF media (Stemgent) until 40% to 50% confluent. iPSCs were transfected in feeder-free media using 1 to 2 µg Puro-T2A-Cre-GFP plasmid DNA with Lipofectamine LTX (Life Technologies) and Lipofectamine PLUS reagent according to the manufacturer’s protocol. Puromycin selection (2-3 µg/mL) was performed for 2 days (starting either 4 hours after transfection or the next day), after which MEFs were added to wells to enhance iPSC colony growth; cells were thereafter cultured in standard iPSC media. Upon sufficient expansion, iPSCs were detached from MEFs by treatment with 1 mg/mL dispase, DNA was extracted, and TaqMan real-time PCR was performed to confirm removal of the loxP-flanked STEMCCA-loxP vector (as described above).
qPCR for gene expression
RNA was isolated by EZ1 RNA Tissue Mini Kit (QIAGEN) from iPSC colonies (detached from MEFs using 1 mg/mL dispase). cDNA was generated from 1 μg total RNA using the SuperScript First-Strand Synthesis System for RT-PCR Kit (Life Technologies). Real-time PCR of pluripotent markers NANOG, OCT4, TERT, and internal control B2M (TaqMan expression assays; Life Technologies) were tested. (Microarray Gene Expression Omnibus accession number: GSE40790.)
iPSC fingerprinting, karyotyping, and gene rearrangement analyses
The identity and origin of iPSC lines were confirmed by microsatellite analysis (Applied Biosystems AmpF? STR COfiler) compared with DNA from the donor’s PBMCs. G-band karyotype analyses of iPSCs were performed commercially (WiCell Research Institute). PCR-based TCRβ, TCRγ, and IgH gene rearrangement analyses of iPSCs (InVivoScribe Technologies) were performed according to the manufacturer’s guidelines.
Additional cell lines
IMR-90 human lung fibroblasts were obtained from ATCC. H1 and H9 human embryonic stem cells (ESCs), used simply as a source of ESC DNA for methylation site pattern analysis comparison with the iPSCs, were obtained from WiCell Research Institute.
Genome-wide analysis of CpG DNA methylation
DNA was isolated using DNeasy blood and tissue kit (QIAGEN). Then 500 ng genomic DNA (picoGreen-quantified) was bisulfite-converted (EZ DNA methylation kit; Zymo Research) and analyzed using Infinium HumanMethylation450 BeadChip (Illumina). SAS and JMP Genomics software (SAS Institute) were used for statistical analysis.
Results
Efficiency of generation of iPSC clones from G-CSF mobilized, purified CD34+ HSCs
We first assessed the efficiency of reprogramming of G-CSF mobilized, purified CD34+-enriched HSCs using STEMCCA-loxP transduction at an MOI of 2. Starting from an 80% to 95% pure population of approximately 20 000 to 50 000 mobilized CD34+ HSCs obtained as described in “Methods,” we generated 4, 10, and 4 iPSC clones, respectively, from 1 healthy donor and 2 patients (Table 1 top section). This set a minimum target of 20 mL of peripheral blood that would be needed with STEMCCA-loxP reprogramming to obtain the number of starting cells necessary to reproducibly generate iPSC clones from the approximately 1400 nonmobilized CD34+ known to be present in each milliliter of peripheral blood.13
List of patients or healthy subjects from whom iPSCs were generated
| Healthy donor or patient with: . | Age (y) . | CD34+ cell purification . | Starting CD34+ cell or blood amount . | TRA-1-60+ colonies . | Number Cre-excised . | Number assessed for 3-germ layer . |
|---|---|---|---|---|---|---|
| STEMCCA-loxP generated iPSC from apheresis collected G-CSF mobilized peripheral blood CD34+ cells | ||||||
| Healthy (iNC-01) | 44 | MACS | 20 000 cells | 4 | 1 | 1 |
| X-CGD (iGP91-07) | 21 | MACS | 50 000 cells | 10 | 1 | 1 |
| X-SCID (iSCID-01) | 17 | Dynal | 50 000 cells | 4 | 1 | 0 |
| STEMCCA-loxP generated iPSCs from nonmobilized peripheral blood CD34+cells | ||||||
| Healthy (iNC-02) | 42 | Dynal | 20 mL | 5 | 1 | 0 |
| Healthy (iNC-03) | 46 | Dynal | 20 mL | 5 | 1 | 0 |
| Healthy (iNC-04) | 52 | MACS | 14 mL | 7 | 1 | 0 |
| Healthy (iNC-05) | 38 | MACS | 28 mL | 3 | 1 | 0 |
| X-CGD (iGP91-01) | 42 | Dynal | 20 mL | 6 | 1 | 1 |
| X-CGD (iGP91-02) | 39 | Dynal | 20 mL | 2 | 1 | 1 |
| X-CGD (iGP91-03) | 36 | MACS | 20 mL | 2 | 2 | 2 |
| X-CGD (iGP91-04) | 30 | MACS | 18 mL | 1 | 1 | 1 |
| X-CGD (iGP91-05) | 12 | MACS | 20 mL | 5 | 0 | 0 |
| AR-CGD (iP22-01) | 19 | MACS | 20 mL | 10 | 1 | 0 |
| AR-CGD (iP47-01) | 37 | Dynal | 20 mL | 5 | 1 | 1 |
| AR-CGD (iP47-02) | 38 | Dynal | 20 mL | 9 | 1 | 1 |
| AR-CGD (iP47-03) | 30 | Dynal | 20 mL | 6 | 1 | 1 |
| AR-CGD (iP47-04) | 23 | MACS | 30 mL | 4 | 0 | 0 |
| AR-CGC (iP67-01) | 51 | MACS | 25 mL | 5 | 1 | 0 |
| X-SCID (iSCID-02) | 17 | MACS | 10 mL | 4 | 1 | 0 |
| X-SCID (iSCID-03) | 19 | MACS | 30 mL | 10 | 1 | 0 |
| X-CGD (iGP91-06) | 16 | MACS | Cryopreserved PBMC 20 mL | 3 | 1 | 0 |
| X-CGD (iGP91-08) | 52 | MACS | Cryopreserved PBMC 20 mL | 6 | 0 | 0 |
| AR-CGD (iP67-02) | 5 | MACS | Cryopreserved PBMC 10 mL | 1 | 1 | 0 |
| Sendai vectors generated iPSCs from nonmobilized peripheral blood CD34+ cells | ||||||
| Healthy (iNC-04-s) | 52 | MACS | 6 mL | >120 (8) | n/a | 2 |
| Healthy (iNC-06-s) | 51 | MACS | 5 mL | 3 (3) | n/a | 0 |
| X-CGD (iGP91-09-s) | 34 | MACS | 9 mL | 10 (5) | n/a | 0 |
| X-CGD (iGP91-10-s) | 22 | MACS | 4 mL | 274 (12) | n/a | 0 |
| X-CGD (iGP91-11-s) | 25 | MACS | 9 mL | 17 (8) | n/a | 0 |
| X-CGD (iGP91-12-s) | 2 | MACS | 2.5 mL | 408 (24) | n/a | 0 |
| Healthy donor or patient with: . | Age (y) . | CD34+ cell purification . | Starting CD34+ cell or blood amount . | TRA-1-60+ colonies . | Number Cre-excised . | Number assessed for 3-germ layer . |
|---|---|---|---|---|---|---|
| STEMCCA-loxP generated iPSC from apheresis collected G-CSF mobilized peripheral blood CD34+ cells | ||||||
| Healthy (iNC-01) | 44 | MACS | 20 000 cells | 4 | 1 | 1 |
| X-CGD (iGP91-07) | 21 | MACS | 50 000 cells | 10 | 1 | 1 |
| X-SCID (iSCID-01) | 17 | Dynal | 50 000 cells | 4 | 1 | 0 |
| STEMCCA-loxP generated iPSCs from nonmobilized peripheral blood CD34+cells | ||||||
| Healthy (iNC-02) | 42 | Dynal | 20 mL | 5 | 1 | 0 |
| Healthy (iNC-03) | 46 | Dynal | 20 mL | 5 | 1 | 0 |
| Healthy (iNC-04) | 52 | MACS | 14 mL | 7 | 1 | 0 |
| Healthy (iNC-05) | 38 | MACS | 28 mL | 3 | 1 | 0 |
| X-CGD (iGP91-01) | 42 | Dynal | 20 mL | 6 | 1 | 1 |
| X-CGD (iGP91-02) | 39 | Dynal | 20 mL | 2 | 1 | 1 |
| X-CGD (iGP91-03) | 36 | MACS | 20 mL | 2 | 2 | 2 |
| X-CGD (iGP91-04) | 30 | MACS | 18 mL | 1 | 1 | 1 |
| X-CGD (iGP91-05) | 12 | MACS | 20 mL | 5 | 0 | 0 |
| AR-CGD (iP22-01) | 19 | MACS | 20 mL | 10 | 1 | 0 |
| AR-CGD (iP47-01) | 37 | Dynal | 20 mL | 5 | 1 | 1 |
| AR-CGD (iP47-02) | 38 | Dynal | 20 mL | 9 | 1 | 1 |
| AR-CGD (iP47-03) | 30 | Dynal | 20 mL | 6 | 1 | 1 |
| AR-CGD (iP47-04) | 23 | MACS | 30 mL | 4 | 0 | 0 |
| AR-CGC (iP67-01) | 51 | MACS | 25 mL | 5 | 1 | 0 |
| X-SCID (iSCID-02) | 17 | MACS | 10 mL | 4 | 1 | 0 |
| X-SCID (iSCID-03) | 19 | MACS | 30 mL | 10 | 1 | 0 |
| X-CGD (iGP91-06) | 16 | MACS | Cryopreserved PBMC 20 mL | 3 | 1 | 0 |
| X-CGD (iGP91-08) | 52 | MACS | Cryopreserved PBMC 20 mL | 6 | 0 | 0 |
| AR-CGD (iP67-02) | 5 | MACS | Cryopreserved PBMC 10 mL | 1 | 1 | 0 |
| Sendai vectors generated iPSCs from nonmobilized peripheral blood CD34+ cells | ||||||
| Healthy (iNC-04-s) | 52 | MACS | 6 mL | >120 (8) | n/a | 2 |
| Healthy (iNC-06-s) | 51 | MACS | 5 mL | 3 (3) | n/a | 0 |
| X-CGD (iGP91-09-s) | 34 | MACS | 9 mL | 10 (5) | n/a | 0 |
| X-CGD (iGP91-10-s) | 22 | MACS | 4 mL | 274 (12) | n/a | 0 |
| X-CGD (iGP91-11-s) | 25 | MACS | 9 mL | 17 (8) | n/a | 0 |
| X-CGD (iGP91-12-s) | 2 | MACS | 2.5 mL | 408 (24) | n/a | 0 |
Shown in each column starting from the left column for each iPSC derivation are: (1) Whether the donor is a healthy donor or has a type of CGD or X-SCID with iPSC line source name designation in parenthesis, and where all subject blood cell sources are unique, with one exception being that the same blood sample from a single healthy subject iHC-04 was split to generate iPSC using STEMCCA-loxP (designated iHC-04) or using Sendai virus (designated iHC-04-s). (2) Donor age. (3) Whether MACS or Dynal was used for CD34+ cell purification. (4) Starting CD34+ cell number or blood volume as indicated. (5) Number of TRA-1-60 vital-stain positive iPSC colonies arising in the culture that were picked, expanded, and cryopreserved (for the Sendai reprogramming group, the estimated number arising in the culture is shown, but the number in parenthesis are the actual number of clones picked, successfully expanded, and cryopreserved). (6) Indicates whether “1” or “2” colonies from that group had completed verification of Cre-excision of single-copy STEMCCA-loxP insert, verification of source identity, continued expression of pluripotency markers, expansion, and cryopreservation (“0” indicates reprogramming source where Cre-excision of a clone with full-line qualification is still in progress). For the Sendai group, the issue of whether Cre-excision was done is not applicable. Note that identity testing for the Cre-excised lines from the 3 X-SCID patients included demonstration that the patient’s known mutation of IL2RG is present in the iPSC line. (7) This column indicates if “1” or “2” of these lines has fully completed either embryoid body three germ layer histology analysis and/or teratoma formation testing in NSG mice (“0” indicates this testing is still in progress); for the STEMCCA-loxP group, all those indicated in column 7 are fully characterized Cre-excised lines.
AR-CGD, autosomal recessive p22phox (due to CYBA mutation), p47phox (due to NCF1 mutation), or p67phox (due to NCF2 mutation) deficient CGD; Dynal, Dynal anti-CD34 magnetic immunobead system; MACS, Miltenyi anti-CD34 magnetic immunobead system; n/a, not applicable; PBMC, peripheral blood mononuclear cells; X-CGD, X-linked gp91phox deficient chronic granulomatous disease (due to CYBB mutation); X-SCID, X-linked severe combined immune deficiency (due to IL2RG mutation).
Efficiency of generation of iPSC clones from nonmobilized purified CD34+-enriched HSCs
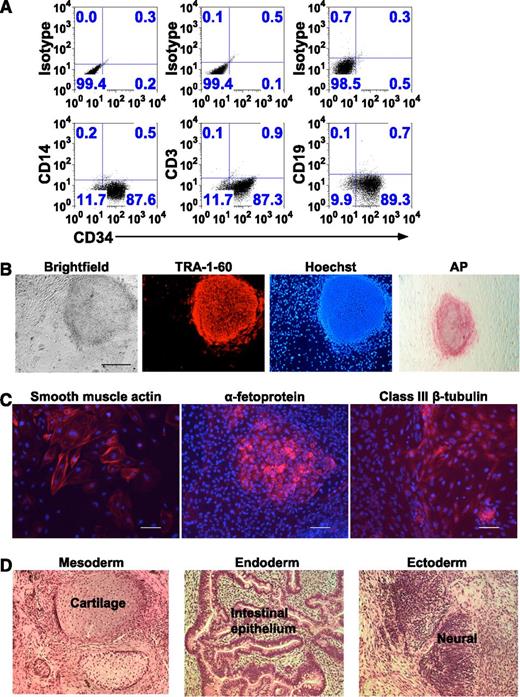
In 8 independent experiments, 20 mL fresh peripheral blood yielded approximately 20 000-50 000 nonmobilized CD34+ HSCs at 80% ± 9% purity (n = 8; Figure 1A) using the separation/purification procedures followed by the 3- to 4-day expansion in HSC media culture. Based on this result, we obtained 10 to 30 mL fresh peripheral blood from 4 healthy donors and from 13 patients as indicated in Table 1, and applied the separation/purification procedures, 3 to 4 days expansion in HSC media culture, transduction with STEMCCA-loxP at an MOI of 2, followed by growth in conditions to generate iPSC clones as described in “Methods.” The iPSC colonies emerged at 9 to 14 days posttransduction and were picked at 18 to 35 days based on morphology and staining for TRA-1-60 pluripotency marker (Figure 1B). Notably, >95% of viable colonies obtained were TRA-1-60+, demonstrating the completeness of STEMCCA-loxP reprogramming of CD34+-enriched HSCs. In these 17 independent iPSC colony generation experiments from fresh peripheral blood from 17 different nonmobilized subjects, the yield of iPSC colonies ranged from 1 to 10 with a median of 5. When the data are normalized to the amount of starting peripheral blood, the iPSC colony yield is 5.3 ± 2.8 iPSC colonies (n = 17) per 20 mL nonmobilized peripheral blood. Because we maintain a large long-term repository of cryopreserved PBMC from patients followed under protocol in our program of study of patients with primary immune deficiencies, it would be useful to know if these procedures for generation of iPSC clones worked equally well on cryopreserved PBMC prepared from approximately 10 to 20 mL peripheral blood. As shown in Table 1, the yield from cryopreserved PBMC samples from 3 chronic granulomatous disease (CGD) patients was 1 to 6 iPSC colonies.
Generation and characterization of iPSCs from nonmobilized CD34+-enriched HSCs isolated from peripheral blood. (A) Flow cytometry dot plot analyses of 1 representative experiment of 8 in which CD34+-enriched HSCs were isolated from 20 mL peripheral blood and placed in culture in HSC media for 4 days, but numerical data for all 8 experiments is provided in the legend text below. The resultant cells in the culture were labeled with anti-CD34, anti-CD14, anti-CD3, and anti-CD19 antibodies. The upper 3 panels show the isotype antibody controls for setting the gating used in each analysis indicated in the lower panels. Shown on the horizontal axis of the lower panels is the expression of CD34 by cells in this representative culture. Shown on the vertical axis of the 3 lower panels starting from the left is the expression of CD14, CD3, and CD19, respectively. In this representative culture, approximately 88% of cells are CD34+. For each of the experiments from 8 donors, the results were 75%D, 84%D, 91%M, 76%M, 88%M, and 69%M CD34+, respectively (where D is Dynal and M is Miltenyi bead isolation). The mean for the group is 80% ± 9% CD34+, with less than 1% of the CD34 dim or negative cells expressing any mature lineage markers in any of the 8 experiments. The lack of lineage markers on the remaining cells in the culture indicates that even the CD34+ dim and negative cells likely derive from proliferation and early loss of CD34 expression by cells that were CD34+ when placed in culture. In this representative experiment, there were no mature monocytes, T lymphocytes, or B lymphocytes detected in the upper left quadrant of the lower panels (CD34 negative/lineage marker strongly positive), and for the entire group of 8 independent experiments, mature monocytes, T lymphocytes, or B lymphocytes represented significantly less than 1% of the population in every case. (B) Microscopy detection of TRA-1-60 and AP pluripotency markers in a representative iPSC line generated by reprogramming of ex vivo cultured nonmobilized CD34+ cells derived from peripheral blood. Shown from left to right is a bright-field phase contrast image of a typical iPSC colony and the same colony immune-fluorescent stained for TRA-1-60 or Hoechst nucleus marker as viewed in fluorescence imaging. In the right-most panel is another colony from the same iPSC line stained for AP as viewed in bright-field microscopy. These images show that iPSC colonies exhibit characteristic ESC-like morphology and express pluripotency markers (bar = 500 µm). For bright-field microscopy, TRA-1-60, and Hoescht, we used a Nikon Eclipse Ti microscope (Melville, NY), Nikon Digital Sight Ds-Qi MC camera, NIS Elements BR 3.10 software; ×4 objective/0.13 numerical aperture, and fluorochromes Alexa-555 and Hoechst. For AP, we used a AMG AMS-MV64 LCD viewing microscope/camera (AMG; Bothel, WA) and Micron 2.0.0 software. (C) Fluorescence microscopy imaging detection of germ layer differentiation markers in EBs generated from a representative iPSC line reprogrammed from ex vivo cultured nonmobilized CD34+ cells derived from peripheral blood. From the left, respectively, the images show that differentiated cells stain positive (red) for smooth muscle actin (mesoderm), α-fetoprotein (endoderm), or class III β-tubulin (ectoderm). Cell nuclei were stained with Hoechst dye in blue (bar = 100 µm). The same Nikon microscopy/camera/software and fluorochromes described above (B) were used: ×10 objective/0.30 numerical aperture. (D) Bright-field microscopy H&E stain demonstration of the differentiation of all 3 germ layers in teratomas arising in immunodeficient NSG mice injected with a representative iPSC line reprogrammed from ex vivo cultured nonmobilized CD34+ cells derived from peripheral blood. From the left, respectively, are sections from the same H&E-stained teratoma containing cartilage (mesoderm), intestinal epithelium (endoderm), and neural tissues (ectoderm). We used a Zeiss Axio Imager.Z1 microscope (Thornwood, NY), a PixeLINK PL-A662 camera with PixeLINK Capture SE software (Ottowa, Ontario), and a ×20 objective/0.75 numerical aperture.
Generation and characterization of iPSCs from nonmobilized CD34+-enriched HSCs isolated from peripheral blood. (A) Flow cytometry dot plot analyses of 1 representative experiment of 8 in which CD34+-enriched HSCs were isolated from 20 mL peripheral blood and placed in culture in HSC media for 4 days, but numerical data for all 8 experiments is provided in the legend text below. The resultant cells in the culture were labeled with anti-CD34, anti-CD14, anti-CD3, and anti-CD19 antibodies. The upper 3 panels show the isotype antibody controls for setting the gating used in each analysis indicated in the lower panels. Shown on the horizontal axis of the lower panels is the expression of CD34 by cells in this representative culture. Shown on the vertical axis of the 3 lower panels starting from the left is the expression of CD14, CD3, and CD19, respectively. In this representative culture, approximately 88% of cells are CD34+. For each of the experiments from 8 donors, the results were 75%D, 84%D, 91%M, 76%M, 88%M, and 69%M CD34+, respectively (where D is Dynal and M is Miltenyi bead isolation). The mean for the group is 80% ± 9% CD34+, with less than 1% of the CD34 dim or negative cells expressing any mature lineage markers in any of the 8 experiments. The lack of lineage markers on the remaining cells in the culture indicates that even the CD34+ dim and negative cells likely derive from proliferation and early loss of CD34 expression by cells that were CD34+ when placed in culture. In this representative experiment, there were no mature monocytes, T lymphocytes, or B lymphocytes detected in the upper left quadrant of the lower panels (CD34 negative/lineage marker strongly positive), and for the entire group of 8 independent experiments, mature monocytes, T lymphocytes, or B lymphocytes represented significantly less than 1% of the population in every case. (B) Microscopy detection of TRA-1-60 and AP pluripotency markers in a representative iPSC line generated by reprogramming of ex vivo cultured nonmobilized CD34+ cells derived from peripheral blood. Shown from left to right is a bright-field phase contrast image of a typical iPSC colony and the same colony immune-fluorescent stained for TRA-1-60 or Hoechst nucleus marker as viewed in fluorescence imaging. In the right-most panel is another colony from the same iPSC line stained for AP as viewed in bright-field microscopy. These images show that iPSC colonies exhibit characteristic ESC-like morphology and express pluripotency markers (bar = 500 µm). For bright-field microscopy, TRA-1-60, and Hoescht, we used a Nikon Eclipse Ti microscope (Melville, NY), Nikon Digital Sight Ds-Qi MC camera, NIS Elements BR 3.10 software; ×4 objective/0.13 numerical aperture, and fluorochromes Alexa-555 and Hoechst. For AP, we used a AMG AMS-MV64 LCD viewing microscope/camera (AMG; Bothel, WA) and Micron 2.0.0 software. (C) Fluorescence microscopy imaging detection of germ layer differentiation markers in EBs generated from a representative iPSC line reprogrammed from ex vivo cultured nonmobilized CD34+ cells derived from peripheral blood. From the left, respectively, the images show that differentiated cells stain positive (red) for smooth muscle actin (mesoderm), α-fetoprotein (endoderm), or class III β-tubulin (ectoderm). Cell nuclei were stained with Hoechst dye in blue (bar = 100 µm). The same Nikon microscopy/camera/software and fluorochromes described above (B) were used: ×10 objective/0.30 numerical aperture. (D) Bright-field microscopy H&E stain demonstration of the differentiation of all 3 germ layers in teratomas arising in immunodeficient NSG mice injected with a representative iPSC line reprogrammed from ex vivo cultured nonmobilized CD34+ cells derived from peripheral blood. From the left, respectively, are sections from the same H&E-stained teratoma containing cartilage (mesoderm), intestinal epithelium (endoderm), and neural tissues (ectoderm). We used a Zeiss Axio Imager.Z1 microscope (Thornwood, NY), a PixeLINK PL-A662 camera with PixeLINK Capture SE software (Ottowa, Ontario), and a ×20 objective/0.75 numerical aperture.
To further explore alternative methods of reprogramming CD34+-enriched HSCs, we tested the combination of four CytoTune Sendai virus reprogramming vectors separately encoding Oct4, Sox2, c-Myc, and Klf4 using cultured CD34+ HSC enriched from 2.5 to 9 mL peripheral blood obtained from 2 healthy donors and 4 patients with CGD. Using this vector system there were widely varying efficiencies of colony yield, and the colonies arising from the peripheral blood–derived CD34+ HSC were variably reprogrammed such that in the 6 independent reprogramming experiments with blood cells from 6 donors, 10% to 80% of colonies that arose were TRA-1-60+. The actual yield of TRA-1-60 colonies arising ranged from as many as >500 to as few as 6 fully reprogrammed iPSC colonies per 10 mL blood (Table 1). These studies demonstrate the suitability of CD34+-enriched HSCs as targets for iPSC reprogramming using nonintegrating viral vectors. The Sendai reprogramming system provides an alternative method resulting in increased colony numbers, with a trade-off of variable yield and increased effort required to screen for fully reprogrammed iPSCs.
Initial characterization of iPSC colonies generated from nonmobilized CD34+-enriched HSCs
In preliminary experiments using different MOI for transduction reprogramming with STEMCCA-loxP, the yield of iPSC colonies generated from CD34+-enriched HSCs increased as MOI was increased, but the standard MOI of 2 used for the studies shown in this report was chosen because the efficiency of transduction was high enough so that the yield of iPSC was adequate but low enough so that most iPSC clones had a copy number of 1. When copy number was determined for the iPSC colonies generated as reported in this study, approximately 95% of clones had a single insert copy of STEMCCA-loxP per cell and the remainder had 2 insert copies per cell. Putative iPSC colonies were analyzed for additional pluripotency markers including AP activity (Figure 1B). iPSCs containing a single vector insert were capable of differentiating into all 3 germ layers in vitro in EB differentiation assays (Figure 1C) and in vivo in teratoma assays (Figure 1D). Similar measures of pluripotency were performed with Sendai reprogrammed iPSC with similar results (not shown).
Cre-mediated excision of STEMCCA-loxP vector insert and characterization of resultant clones
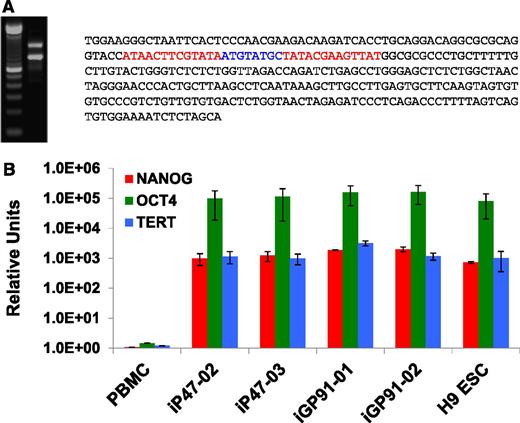
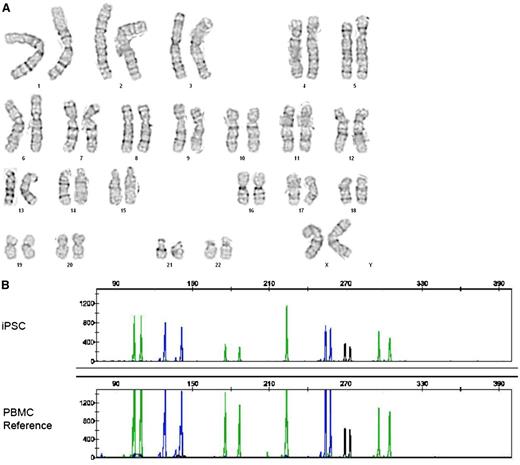
We excised STEMCCA-loxP vector from 1 to 2 single-copy iPSC lines from each donor indicated in Table 1 by transiently expressing Cre recombinase by transfection with the Puro-T2A-Cre-GFP plasmid as indicated in “Methods.” Puromycin selection was performed for 2 days after transfection and colonies were expanded thereafter in the absence of puromycin. GFP expression typically disappeared at 2 to 3 days posttransfection and expanded colonies were negative for GFP expression, confirming transient retention of the Puro-T2A-Cre-GFP plasmid. STEMCCA-loxP excision was confirmed by qPCR. LAM-PCR17 was performed on some of these clones to determine the integration site (Table 2), allowing characterization of the residual vector footprint after excision (Figure 2A). All STEMCCA-loxP–excised iPSC colonies assessed thus far retained pluripotency markers and capacity for 3 germ layer formation in EBs and in teratomas (Table 1). RNA analysis confirmed continued expression of pluripotent markers NANOG, OCT4, and TERT in iPSC clones from which the reprogramming vector had been excised, comparable to levels expressed in H9 ESCs (Figure 2B). After excision, iPSCs also exhibited normal karyotype (Figure 3A) and DNA fingerprinting confirmed the identity and origin of iPSCs by comparison with patient PBMCs (Figure 3B). Mutation sequencing analysis also confirmed that iPSC lines matched the patient (data not shown). Of particular note, the 3 X-SCID patients who had received haploidentical bone marrow grafts as infants retained only unilineage donor CD3 lymphocyte engraftment at time of blood draw. All iPSC lines derived from these X-SCID patients contained the subject’s mutation in IL2RG; 879C>T (iSCID-01), 529T>C (iSCID-02), and 447delA (iSCID-03).
List of LAM-PCR integration analysis sites for STEMCCA-loxP
| iPSC line . | Chromosome . | bp position . | Gene . |
|---|---|---|---|
| iNC-01 | Chr 17 | 76054451 | TNRC6C (intron) |
| iGP91-02 | Chr 6 | 147435589 | ENSG00000233452 |
| iGP91-03-1 | Chr 1 | 44312073 | ST3GAL3 (intron) |
| iGP91-03-2 | Chr 22 | 21115470 | PI4KA (intron) |
| iGP91-04 | Chr 17 | 3618824 | ITGAE (intron) |
| iP47-01 | Chr 3 | 112952023 | BOC (intron) |
| iP47-02 | Chr 1 | 39677115 | MACF1 (intron) |
| iPSC line . | Chromosome . | bp position . | Gene . |
|---|---|---|---|
| iNC-01 | Chr 17 | 76054451 | TNRC6C (intron) |
| iGP91-02 | Chr 6 | 147435589 | ENSG00000233452 |
| iGP91-03-1 | Chr 1 | 44312073 | ST3GAL3 (intron) |
| iGP91-03-2 | Chr 22 | 21115470 | PI4KA (intron) |
| iGP91-04 | Chr 17 | 3618824 | ITGAE (intron) |
| iP47-01 | Chr 3 | 112952023 | BOC (intron) |
| iP47-02 | Chr 1 | 39677115 | MACF1 (intron) |
Shown are the single-copy STEMCCA-loxP integration sites in 7 independent iPSC lines determined by LAM-PCR before Cre-excision and verified by flanking genome primer PCR facilitated sequencing after Cre-excision. The chromosome, base pair position within the chromosome, and gene location in which insertion occurred (when indicated) were determined using ENSEMBL release 67, Homo sapiens high coverage assembly GRCh37 (GCA_000001405.6) from the Genome Reference Consortium. The 303-bp residual STEMCCA-loxP footprint sequence at the indicated genomic site was identical for all 7 clones analyzed by LAM-PCR and postexcision sequencing, as shown in Figure 2. The line source name is as designated in Table 1. Note that in the case of donor source iGP91-03, 2 clones (iGP91-03-1 and iGP91-03-2) arising in the same culture from that reprogramming experiment were analyzed.
Chr, chromosome.
Analysis of Cre-mediated excision of the reprogramming STEMCCA-loxP vector genomic insert from iPSCs and demonstration of continued expression of native pluripotency genes following excision. (A) Shown is the PCR (left) and sequencing (right) analyses of the residual genomic footprint left after Cre-mediated loxP excision from single-copy reprogramming STEMCCA-loxP vector genomic insert from one of the iPSC lines reprogrammed from nonmobilized CD34+ cells derived from peripheral blood. Following Cre-GFP transfection, 20% to 75% of all colonies from all donors tested had the vector removed and no colonies retained GFP expression. For this representative cell line, LAM-PCR analysis was used to determine the genomic site of the single-copy insert of STEMCCA-loxP. Primers were designed that matched human genomic sequences at least 300 to 500 bp flanking the insert and were used to generate standard PCR products from genomic DNA as shown in this agarose gel image on the left. The left lane in the gel is the 100-bp sizing ladder used to estimate the sizes of the PCR products in this example. The right lane shows that 2 bands approximately 300-bp different in size are generated from the primer pair, where the lower band is derived from the unaltered allele’s native genomic sequence of approximately 800 bp that separates the primer binding sites, while the upper band is approximately 300-bp larger derived from the allele that contains the same intervening human sequence plus the residual vector footprint following Cre-mediated loxP excision. The footprint sequence was determined by cloning and sequencing this upper band PCR product. Shown on the right is the entire 303-bp residual STEMCCA-loxP footprint sequence consisting of 1 complete vector LTR sequence (black) with the 5′ and 3′ portions of that LTR sequence separated by a single intervening loxP site derived from the recombination excision event. The 2 palindromic ends of the loxP are shown in red while the nonpalindromic middle section of the loxP sequence is shown in blue. (B) Bar graphs showing quantitative reverse-transcription PCR detection of expression of mRNAs encoding for pluripotent markers NANOG (red), OCT4 (green), and TERT (blue) in several representative CD34+ cell-derived iPSCs from which STEMCCA-loxP had been excised with Cre. Expression in the Cre-excised iPSC lines are compared with H9 ESCs with the TaqMan-based expression data normalized to expression of human B2M (β-2-microglobulin) as an endogenous control and expressed in relative units for comparison. PBMCs were used as the calibrator control for expression comparisons. All samples were analyzed in quadruplicate.
Analysis of Cre-mediated excision of the reprogramming STEMCCA-loxP vector genomic insert from iPSCs and demonstration of continued expression of native pluripotency genes following excision. (A) Shown is the PCR (left) and sequencing (right) analyses of the residual genomic footprint left after Cre-mediated loxP excision from single-copy reprogramming STEMCCA-loxP vector genomic insert from one of the iPSC lines reprogrammed from nonmobilized CD34+ cells derived from peripheral blood. Following Cre-GFP transfection, 20% to 75% of all colonies from all donors tested had the vector removed and no colonies retained GFP expression. For this representative cell line, LAM-PCR analysis was used to determine the genomic site of the single-copy insert of STEMCCA-loxP. Primers were designed that matched human genomic sequences at least 300 to 500 bp flanking the insert and were used to generate standard PCR products from genomic DNA as shown in this agarose gel image on the left. The left lane in the gel is the 100-bp sizing ladder used to estimate the sizes of the PCR products in this example. The right lane shows that 2 bands approximately 300-bp different in size are generated from the primer pair, where the lower band is derived from the unaltered allele’s native genomic sequence of approximately 800 bp that separates the primer binding sites, while the upper band is approximately 300-bp larger derived from the allele that contains the same intervening human sequence plus the residual vector footprint following Cre-mediated loxP excision. The footprint sequence was determined by cloning and sequencing this upper band PCR product. Shown on the right is the entire 303-bp residual STEMCCA-loxP footprint sequence consisting of 1 complete vector LTR sequence (black) with the 5′ and 3′ portions of that LTR sequence separated by a single intervening loxP site derived from the recombination excision event. The 2 palindromic ends of the loxP are shown in red while the nonpalindromic middle section of the loxP sequence is shown in blue. (B) Bar graphs showing quantitative reverse-transcription PCR detection of expression of mRNAs encoding for pluripotent markers NANOG (red), OCT4 (green), and TERT (blue) in several representative CD34+ cell-derived iPSCs from which STEMCCA-loxP had been excised with Cre. Expression in the Cre-excised iPSC lines are compared with H9 ESCs with the TaqMan-based expression data normalized to expression of human B2M (β-2-microglobulin) as an endogenous control and expressed in relative units for comparison. PBMCs were used as the calibrator control for expression comparisons. All samples were analyzed in quadruplicate.
G-band karyotype analysis and DNA fingerprint analysis of iPSCs. (A) Cytogenetic analysis of a representative nonmobilized CD34+-cell–derived iPSC line after Cre-mediated excision of STEMCCA-loxP, demonstrating a normal karyotype. (B) The identity and origin of that iPSC line is confirmed by DNA microsatellite analysis at 7 loci in comparison with PBMCs derived from that patient. DNA samples were PCR amplified using the Applied Biosystems AmpF? STR COfiler kit and the PCR products were analyzed with an ABI 3130XL Genetic Analyzer using GeneMapper software.
G-band karyotype analysis and DNA fingerprint analysis of iPSCs. (A) Cytogenetic analysis of a representative nonmobilized CD34+-cell–derived iPSC line after Cre-mediated excision of STEMCCA-loxP, demonstrating a normal karyotype. (B) The identity and origin of that iPSC line is confirmed by DNA microsatellite analysis at 7 loci in comparison with PBMCs derived from that patient. DNA samples were PCR amplified using the Applied Biosystems AmpF? STR COfiler kit and the PCR products were analyzed with an ABI 3130XL Genetic Analyzer using GeneMapper software.
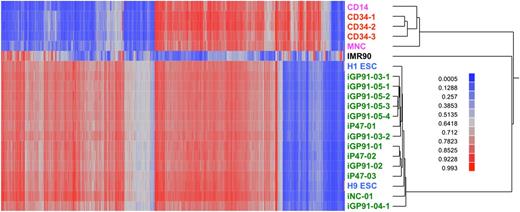
Additional characterization included comparison of genomic methylation pattern of 13 Cre-excised iPSC lines with that of human ESCs and adult somatic cells. As shown in Figure 4, our nonmobilized CD34+-enriched HSC-derived iPSCs exhibit DNA methylation profiles that very closely match that of reference human ESC lines and are distinct from DNA methylation profiles of CD34+-enriched HSCs, CD14+ monocytes, a mixture of PBMCs, or fibroblasts. We have not yet completed methylation pattern analysis of Sendai-reprogrammed iPSC clones.
Differential DNA methylation of human ESCs, iPSCs, and primary cells of various sources. These data show that iPSCs generated using our method from peripheral blood nonmobilized CD34+ cells have a general pattern of CpG methylation almost indistinguishable from ESCs and dramatically different from primary hematopoietic cells (CD14 monocytes, 3 separate samples of CD34+ cells, or mixed PBMCs) or the IMR-90 primary fibroblast line. The heat map encodes β values (red, methylated; blue, not methylated) of 2404 autosomal CpG sites (columns) and DNA samples (rows). The Illumina Infinium assay probed 485K methylation sites. Analysis of variance was used to test for a difference in average methylation level (β) between all pairs of 6 predefined classes of cells: ESCs (blue text), CD34+-derived iPSCs (green text), CD34+ cells (red text), PBMCs and CD14+ cells (pink text), and fibroblasts (black text). The CD34+-derived iPSCs include 13 independent lines from 9 different donors, 5 of which had the STEMCCA-loxP vector excised by Cre recombinase (iNC-01, iGP91-03-1, iGP91-03-2, iGP91-04, and iP47-02). We identified a total of 24 040 sites differentially methylated for at least 1 group comparison at a significance level of P < 1 × 10−12; 10% of those sites were selected at random for the subsequent cluster analysis of individual DNA samples. These methylation data are available under GEO accession number GSE40790.
Differential DNA methylation of human ESCs, iPSCs, and primary cells of various sources. These data show that iPSCs generated using our method from peripheral blood nonmobilized CD34+ cells have a general pattern of CpG methylation almost indistinguishable from ESCs and dramatically different from primary hematopoietic cells (CD14 monocytes, 3 separate samples of CD34+ cells, or mixed PBMCs) or the IMR-90 primary fibroblast line. The heat map encodes β values (red, methylated; blue, not methylated) of 2404 autosomal CpG sites (columns) and DNA samples (rows). The Illumina Infinium assay probed 485K methylation sites. Analysis of variance was used to test for a difference in average methylation level (β) between all pairs of 6 predefined classes of cells: ESCs (blue text), CD34+-derived iPSCs (green text), CD34+ cells (red text), PBMCs and CD14+ cells (pink text), and fibroblasts (black text). The CD34+-derived iPSCs include 13 independent lines from 9 different donors, 5 of which had the STEMCCA-loxP vector excised by Cre recombinase (iNC-01, iGP91-03-1, iGP91-03-2, iGP91-04, and iP47-02). We identified a total of 24 040 sites differentially methylated for at least 1 group comparison at a significance level of P < 1 × 10−12; 10% of those sites were selected at random for the subsequent cluster analysis of individual DNA samples. These methylation data are available under GEO accession number GSE40790.
Demonstration that iPSCs are not derived from lymphocytes or monocytes
We are not the first to report that iPSCs can be generated from PBMCs derived from nonmobilized peripheral blood, but without performing the maneuvers that we stipulate in “Methods” to purify the CD34+ cells and using culture conditions that bias for expansion of CD34+ HSCs, it is not possible to have a high level of certainly from which lineage blood cell the iPSC are derived. In fact, reprogramming of unsorted PBMCs has been demonstrated to result in T-cell–derived iPSCs exhibiting TCR recombination changes in the genome,4,5 and it has been assumed that some of the remainder of such PBMC-derived iPSCs not containing lymphocyte-related genomic recombination changes are derived from monocytes. We performed TCRβ, TCRγ, and IgH gene rearrangement analyses on 22 iPSC lines from 17 different donors (8 lines shown in Figure 5), finding the genome complete and unrecombined at the TCRβ, TCRγ, and IgH gene loci in all cases. Additionally, using our culture and reprogramming conditions in a series of studies using up to 200 000 CD14+ monocytes purified from peripheral blood PBMCs, we have been unable to generate even a single iPSC clone even when STEMCCA-loxP is used for transduction of the monocytes at MOIs up to 10. These observations strongly support the CD34+ HSC origin of our iPSCs derived from nonmobilized peripheral blood using our procedures.
Analyses of TCR and IgH gene rearrangements in iPSC lines. iPSC lines were tested for TCRβ, TCRγ, and IgH recombination by genomic DNA PCR using InvivoScribe Clonality Assays. Eight independent iPSC lines from 7 different donors (lanes 1-8) are shown: iNC-01 (lane 1), iGP91-03-1 (lane 2), iGP91-03-2 (lane 3), iGP91-04 (lane 4), iGP91-05 (lane 5), iGP91-06 (lane 6), iSCID-02 (lane 7), and iSCID-03 (lane 8). InvivoScribe Clonality Assay controls are shown as polyclonal (lane 9) and 2 different clonal controls (lanes 10 and 11) for each set. A 100-bp DNA ladder is also shown (lane 12). PCR products within the valid range of 69 to 129 bp for IgH, 80 to 220 bp for TCRγ, and 170 to 325 bp for TCRβ are shown. The arrow indicates the 100-bp size marker. The data shown are representative of 22 iPSC lines from 17 different donors.
Analyses of TCR and IgH gene rearrangements in iPSC lines. iPSC lines were tested for TCRβ, TCRγ, and IgH recombination by genomic DNA PCR using InvivoScribe Clonality Assays. Eight independent iPSC lines from 7 different donors (lanes 1-8) are shown: iNC-01 (lane 1), iGP91-03-1 (lane 2), iGP91-03-2 (lane 3), iGP91-04 (lane 4), iGP91-05 (lane 5), iGP91-06 (lane 6), iSCID-02 (lane 7), and iSCID-03 (lane 8). InvivoScribe Clonality Assay controls are shown as polyclonal (lane 9) and 2 different clonal controls (lanes 10 and 11) for each set. A 100-bp DNA ladder is also shown (lane 12). PCR products within the valid range of 69 to 129 bp for IgH, 80 to 220 bp for TCRγ, and 170 to 325 bp for TCRβ are shown. The arrow indicates the 100-bp size marker. The data shown are representative of 22 iPSC lines from 17 different donors.
Discussion
In this report, we describe an efficient method of generating iPSCs from a nonmobilized CD34+-cell–enriched population derived from small volumes of peripheral blood using the Cre-excisable polycistronic STEMCCA-loxP lentivector or the combination of nonintegrating Sendai vectors for reprogramming. We also show that this technique is applicable to generation of iPSCs from the nonmobilized CD34+-enriched HSCs present in patient peripheral blood PBMCs held in long-term cryopreservation storage. Using the STEMCCA-loxP reprogramming vector at an MOI of 2, the iPSC colony yield is adequate and reliable from only 20 mL peripheral blood, with nearly all resultant iPSC colonies generated containing only a single copy per cell of the reprogramming vector. Similarly, using nonintegrating Sendai reprogramming, the colony yields of fully reprogrammed iPSC, though variable, are very acceptable using as little as 2.5 to 9 mL peripheral blood. Use of Sendai has the additional advantage of yielding clones with no vector insert. However, the STEMCCA-loxP is very reliable, yield can be increased by increasing MOI, and we provide detailed instructions for a reliable method to efficiently excise the STEMCCA-loxP reprogramming vector from the iPSC clones using transfection-mediated transient Cre expression, yielding reprogramming transgene-free iPSCs that contain only the 303-bp residual genomic footprint consisting of a single recombined promoterless vector LTR and recombined loxP site. The resultant iPSC clones express pluripotency markers, can form teratomas, and have a genomic methylation profile indistinguishable from reference human ESCs.
Nonmobilized peripheral blood obtained by venipuncture provides a convenient source of reprogrammable CD34+ HSCs that does not require bone marrow aspirate, mobilization with G-CSF treatment, apheresis, or even excessively large blood volume. We were able to use as little as 2.5 mL peripheral blood in one of the successful Sendai reprogramming experiments. Although our STEMCCA-loxP method as described was standardized for 20 mL blood, increasing the MOI of STEMCCA-loxP lentivector proportionately increases colony yield, allowing reprogramming from much smaller volumes of peripheral blood or generation of much higher numbers of clones, though more screening is required to identify which iPSC clones have only 1 vector copy integration.
However, there are trade-offs of expense, screening work, and the goals of the iPSC generation that influence which reprogramming system is most desirable for a particular project. The CytoTune Sendai reagents are more expensive, but do result in very large numbers of integrant-free colonies from the CD34+ cells derived from only a few milliliters of blood. However, we find that yields are quite variable and in some experiments more than half of those colonies are not fully reprogrammed.
The methods that we describe are easily adaptable to Good Laboratory Practice or even Good Manufacturing Practice sterile defined closed system preparations, facilitating a future transition to potential clinical applications of iPSCs as such applications are developed. Finally, long-term storage cryopreserved PBMCs can be used as a starting material for generation of iPSC from CD34+-enriched HSCs. Thus, GMP repositories of cryopreserved PBMC from patient cohorts become valuable and adequate sources of starting materials for generation of iPSCs for research and for future therapeutic application.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE40790).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Gustavo Mostoslavsky for the hSTEMCCA-loxP plasmid and Dr Martha Quezado for histological analysis.
All the authors are supported by the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, National Institutes of Health. This work was also supported by an intramural award from the NIH Center for Regenerative Medicine to HLM.
Authorship
Contribution: R.K.M., C.L.S., and H.L.M. wrote the manuscript. C.L.S., J.P., R.K.M., and S.S.D.R. contributed to iPSC reprogramming. C.L.S., L.C., R.K.M., N.L.T., and U.C. performed LAM-PCR/sequencing analysis. C.L.S., G.L., R.K.M., and S.K. characterized iPSCs. F.O.-C. performed microarray sample processing and hybridizations and T.G.M. analyzed DNA methylation.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Harry L. Malech, LHD/NIAID/NIH, Building 10 Room 5-3750, 10 Center Dr, MSC-1456, Bethesda, MD 20892-1456, e-mail: hmalech@nih.gov.
References
Author notes
R.K.M. and C.L.S. contributed equally to this study.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal