Key Points
Outcome prediction in DLBCL.
MYC status in concert with BCL2 and BCL6.
Abstract
MYC rearrangements occur in 5% to 10% of diffuse large B-cell lymphomas (DLBCL) and confer an increased risk to cyclophosphamide, hydroxydaunorubicin, oncovin, and prednisone (CHOP) and rituximab (R)–CHOP treated patients. We investigated the prognostic relevance of MYC-, BCL2- and BCL6-rearrangements and protein expression in a prospective randomized trial. Paraffin-embedded tumor samples from 442 de novo DLBCL treated within the RICOVER study of the German High-Grade Non-Hodgkin Lymphoma Study Group (DSHNHL) were investigated using immunohistochemistry and fluorescence in situ hybridization (FISH) to detect protein expression and breaks of MYC, BCL2, and BCL6. Rearrangements of MYC, BCL2, and BCL6 were detected in 8.8%, 13.5%, and 28.7%, respectively. Protein overexpression of MYC (>40%) was encountered in 31.8% of tumors; 79.6% and 82.8% of tumors expressed BCL2 and BCL6, respectively. MYC translocations, MYChigh, BCL2high, and BCL6low protein expressions were associated with inferior survival. In multivariate Cox regression modeling, protein expression patterns of MYC, BCL2 and BCL6, and MYC rearrangements were predictive of outcome and provided prognostic information independent of the International Prognostic Index (IPI) for overall survival and event-free survival. A combined immunohistochemical or FISH/immunohistochemical score predicts outcome in DLBCL patients independent of the IPI and identifies a subset of 15% of patients with dismal prognosis in the high-risk IPI group following treatment with R-CHOP. Registered at http://www.cancer.gov/clinicaltrials: RICOVER trial of the DSHNHL is NCT 00052936.
Introduction
Diffuse large B-cell lymphomas (DLBCL) constitute a heterogeneous category of aggressive lymphomas.1 Gene expression profiling has helped to stratify DLBCL into biologically meaningful and prognostically relevant subgroups.2,3 The translation of complex gene expression profiling predictors into, eg, immunohistochemistry (IHC)-based assays applicable to paraffin-embedded and formalin-fixed (FFPE) tumor specimens, however—a prerequisite for everyday clinical practice—has been difficult.4,5
Recurrent translocations targeting BCL2, BCL6, and MYC have been described in DLBCL. While different results have been published regarding the impact of BCL2 and BCL6 breaks on prognosis in DLBCL,6-12 MYC translocations confer a worse prognosis in patient cohorts treated with cyclophosphamide, hydroxydaunorubicin, oncovin, and prednisone (CHOP)–like10,11 and CHOP plus rituximab (R-CHOP) regimens.13,14 Of note, patients whose tumors present with “double-hit” features, ie, dual translocations involving both MYC and BCL2 or BCL6 have shown a dismal clinical course.15 However, some questions regarding the deregulation of MYC in DLBCL still exist. First, analyses targeting the importance of MYC in DLBCL have been carried out mostly in population-based cohorts, but rarely within randomized prospective clinical trials.13,14 Second, the prognostic relevance of MYC rearrangements in DLBCL has rarely been studied in the context of concomitant translocations involving BCL2 and BCL6, and expression of these 2 proteins. Third, the high-throughput analysis of fluorescence in situ hybridization (FISH) in the context of large case numbers and—especially—on tissue microarray (TMA) sections has not been convincingly shown to be both reproducible and feasible. Finally, MYC can also be activated and overexpressed by mechanisms other than translocations, eg, by amplifications, mutations, or by microRNA-dependent mechanisms.16-18 Recently, a novel monoclonal antibody has been validated for use in FFPE tissues,19 and nuclear reactivity for this antibody was shown to be predictive of MYC rearrangements.20,21
The aim of the present study was, therefore, to comprehensively assess the prognostic impact of protein expression patterns of MYC, BCL2, and BCL6 in concert with the recurrent chromosomal translocations targeting MYC, BCL2, and BCL6 in DLBCL patients treated in a large, prospective randomized clinical trial.
Material and methods
Study cohort
All cases analyzed in this study were enrolled in the prospective randomized multicenter clinical trial RICOVER-60 of the German High-Grade Lymphoma Study Group (DSHNHL),22 in which patients over 60 years of age with CD20-positive aggressive B-cell lymphomas had been randomly assigned to 6 or 8 cycles of CHOP-14 treatment with or without 8 applications of rituximab. Diagnostic samples from the study patients were reviewed by expert hematopathologists according to the 2008 World Health Organization classification.1 Assembly of tissue microarrays was performed as described previously.4 Altogether, 506 DLBCL specimens were represented on the TMAs. This clinical trial was conducted in accordance with the Helsinki declaration, and the protocol had been approved by the ethics review committee of each participating center.
FISH
Interphase FISH was performed on standard tissue sections according to described protocols.23,24 Vysis LSI®BCL2-, Vysis LSI®BCL6-, and Vysis LSI®MYC-dual color break-apart rearrangement probes (Vysis/Abbott Molecular Diagnostics, Wiesbaden-Delkenheim, Germany) were used. The cutoff levels for the break-apart probes were established by evaluating the split-signal distribution in samples of reactive lymphoid tissues, calculating the mean number of split signals plus 3 times the standard deviation. The cutoff levels were 13%, 12%, and 15% for the BCL2, BCL6, and MYC break-apart probes, respectively. To validate the hybridization efficiency of TMA-spotted samples, we reanalyzed a series of 10 cases with different hybridization results for each probe by performing FISH with BCL2, BCL6, and MYC probes, respectively, on sections of the original FFPE tumor block. Evaluation of the signal distributions in the TMAs was carried out by 2 independent experienced observers in 2 institutions. In case of discordant results between the 2 observers, the hybridization was repeated using sections of the original FFPE tumor block, and a third investigator was involved. Cases judged as not evaluable by 1 of the investigators were excluded from statistical analyses.
Immunohistochemical staining and scoring
Immunohistochemistry with antibodies against CD20, CD10, BCL2, BCL6, IRF4/MUM1, and Ki67 had previously been performed.4 MYC immunohistochemistry was done on TMA sections using the antibody clone Y69 (Epitomics, Burlingame, CA) at a 1:50 dilution and resulted in nuclear staining of variable intensity in the positive cases.19 After applying pressure cooking in citric acid (pH 6.0) for retrieval, detection was accomplished with Dako Advance reagents (K4068). Because of exhaustion of material, MYC was stained in only 310 cases. Immunohistochemistry was evaluated by 2 experienced hematopathologists on a multihead microscope, and results were recorded as the percentage of positive tumor cells in increments of 10% regardless of the staining intensity.25,26 BCL2 and BCL6 stainings had been evaluated previously and had been recorded in steps of 25%. Lymphoma cases were judged as positive or overexpressed for these antigens (BCL2high, BCL6high) if positive in >0% and >25% of cells, respectively (as extensively discussed by Ott et al and Bernd et al.4,27 ). CD10 was judged as positive, if 1% to 100% of tumor cells were positive, and IRF4/MUM1 was rated as positive if >5% of tumor cell nuclei were stained. Determination of these (different) cut points had also been accomplished by taking into account the highest level of reproducibility among different hematopathologists evaluating the initial stainings.4
Statistical evaluation
Event-free survival (EFS) was the main endpoint of the RICOVER-60 trial and is defined as the time from randomization to disease progression, start of salvage treatment, additional (unplanned) treatments, relapse, or death from any cause. Overall survival (OS) was defined as the time from randomization to death of any cause. EFS and OS were estimated according to Kaplan and Meier.28
To determine the cutpoint for statistical analysis discriminating BCL2, BCL6 and MYC staining positive and negative cases, we took into account the sample size within score groups and the effect sizes regarding EFS and OS for different scenarios with different definitions of positivity. The most appropriate cutpoints were 0% (=BCL2low) vs 1% to 100% (=BCL2high) positive cells for BCL2, 0% to 25% (=BCL6low) vs 26% to 100% (=BCL6high) for BCL6 and 0% to 40% (MYClow) vs 41% to 100% (=MYChigh) for MYC. Interestingly, the MYC cutoff point was very similar to that chosen in 2 recent publications by Johnson et al and Green and colleagues.4,25-27
In the univariate outcome analyses, logrank tests were performed. The International Prognostic Index (IPI; ie, age > 60, lactate dehydrogenase [LDH] > normal, Eastern Cooperative Oncology Group (ECOG) > 1, stage III/IV, and extranodal involvement > 1) is the current gold standard for prognostic stratification in DLBCL also in the rituximab era,29 and proportional hazard models for each of the selected parameters were separately adjusted for the factors of the IPI. Relative risks with 95% confidence intervals (CI) and P values are presented. For the correlation of translocation and marker expression with qualitative data (morphology, Germinal Center B-cell type [GCB]/non-GCB subtypes) and for differences regarding patient characteristics, we used χ-square and, if necessary, Fisher’s exact test. Because of the descriptive nature of these comparisons, the P values were not adjusted for multiple comparisons, and the significance level was P = .05. For many of the IHC/translocation markers, we observed a variable number of cases failing to yield reliable staining/FISH results, and accordingly, the latter were excluded from analysis. Thus, the number of evaluable cases varied from one marker to another. Cohens κ statistic was used to assess the concordance between 2 different observers. Statistical analyses were done with SPSS PASW 18 and IBM SPSS Statistics 20.
Results
Clinical and sample characteristics
The clinical features of the DLBCL groups with FISH results are provided in Table 1. Tissue cores from 506 patients were represented on the TMAs, and 442 tumors were amenable to FISH analysis (87.4%). Two hundred seventy-four out of 442 tumors (62.0%) had been classified as centroblastic and 29 of 442 (6.6%) as immunoblastic DLBCL. In 139 (31.5%) lymphomas, no subclassification had been rendered (DLBCL NOS). According to the Hans classifier,30 157 DLBCL (47.0%) were of GCB-type, and 177 (53.0%) of non-GCB-type.4
Clinical and morphological characteristics of DLBCL patients
| All DLBCL patients . | With FISH data (n = 442) . | RICOVER-60 (n = 949) . |
|---|---|---|
| Male | 240 (54.3%) | 511 (53.8%) |
| Female | 202 (45.7%) | 438 (46.2%) |
| Age, median (range) | 68 (61,80) | 69 (61,80) |
| LDH > UNV | 200 (45.2%) | 467 (49.2%) |
| ECOG > 1 | 53 (12.0%) | 134 (14.1%) |
| Stage III/IV | 192 (43.4%) | 462 (48.7%) |
| E > 1 | 64 (14.5%) | 171 (18.0%) |
| IPI score | ||
| 1 | 157 (35.5%) | 300 (31.6%) |
| 2 | 126 (28.5%) | 258 (27.2%) |
| 3 | 104 (23.5%) | 235 (24.8%) |
| 4,5 | 55 (12.4%) | 156 (16.4%) |
| Bulky disease | 154 (34.8%) | 352 (37.1%) |
| B symptoms | 134 (30.3%) | 312 (32.9%) |
| E involvement | 228 (51.6%) | 543 (57.2%) |
| BM involvement | 23 (5.2%) | 52 (5.5%) |
| Centroblastic | 274 (62.0%) | 516 (54.4%) |
| Immunoblastic | 29 (6.6%) | 63 (6.6%) |
| Plasmablastic | 2 (0.5%) | 7 (0.7%) |
| Anaplastic large cell | 8 (1.8%) | 19 (2.0%) |
| T-cell-rich B-cell | 14 (3.2%) | 18 (1.9%) |
| NOS | 111 (25.1%) | 313 (33.0%) |
| Primary mediastinal B-cell | 4 (0.9%) | 13 (1.4%) |
| All DLBCL patients . | With FISH data (n = 442) . | RICOVER-60 (n = 949) . |
|---|---|---|
| Male | 240 (54.3%) | 511 (53.8%) |
| Female | 202 (45.7%) | 438 (46.2%) |
| Age, median (range) | 68 (61,80) | 69 (61,80) |
| LDH > UNV | 200 (45.2%) | 467 (49.2%) |
| ECOG > 1 | 53 (12.0%) | 134 (14.1%) |
| Stage III/IV | 192 (43.4%) | 462 (48.7%) |
| E > 1 | 64 (14.5%) | 171 (18.0%) |
| IPI score | ||
| 1 | 157 (35.5%) | 300 (31.6%) |
| 2 | 126 (28.5%) | 258 (27.2%) |
| 3 | 104 (23.5%) | 235 (24.8%) |
| 4,5 | 55 (12.4%) | 156 (16.4%) |
| Bulky disease | 154 (34.8%) | 352 (37.1%) |
| B symptoms | 134 (30.3%) | 312 (32.9%) |
| E involvement | 228 (51.6%) | 543 (57.2%) |
| BM involvement | 23 (5.2%) | 52 (5.5%) |
| Centroblastic | 274 (62.0%) | 516 (54.4%) |
| Immunoblastic | 29 (6.6%) | 63 (6.6%) |
| Plasmablastic | 2 (0.5%) | 7 (0.7%) |
| Anaplastic large cell | 8 (1.8%) | 19 (2.0%) |
| T-cell-rich B-cell | 14 (3.2%) | 18 (1.9%) |
| NOS | 111 (25.1%) | 313 (33.0%) |
| Primary mediastinal B-cell | 4 (0.9%) | 13 (1.4%) |
UNV, >normal; B, B-cell; E, extranodal sites; and BM, bone marrow.
FISH analyses
Initially, we created a pilot series of 46 B-NHL tumor samples spotted on TMAs and with available classical chromosome banding data (at least for BCL2, BLC6, and MYC translocations). This series was studied by FISH on the TMA using break-apart probes for BCL2, BCL6, MYC, and IGH. In addition, 25 samples were also studied on whole tissue sections. Of these 100 hybridization events, all 87 hybridizations evaluable (87%) showed concordant results on the TMA and the whole tissue sections, while 13 hybridizations (13%) were not evaluable on the TMA. The results obtained by FISH on the TMA showed a correlation of 94% to the chromosome-banding approaches (data not shown). Of 442 DLBCL specimens hybridized, 384 (86.9%), 404 (91.4%), and 407 (92.1%) samples were interpretable by 2 independent observers for the BCL2, BCL6, and MYC break-apart probes used, respectively. Comparison of the hybridization results for BCL2, BCL6, and MYC break-apart probes in 10 TMA-spotted samples and in the corresponding whole-tissue sections from the original FFPE tumor blocks (30 hybridizations) yielded concordant results in all hybridizations covering samples with different translocation events. With regard to the independent evaluation of FISH signal distributions by 2 observers in 2 institutions, highly satisfactory κ-values of 0.92, 0.97, and 0.94 for BCL2, BCL6, and MYC rearrangements, respectively, were achieved. Discordant results between the initial 2 investigators were observed in 6 (6 of 384, 1.6%), 5 (5 of 404, 1.2%), and 4 (4 of 407, 1.0%) samples for BCL2, BCL6, and MYC translocation probes, respectively, which all could be resolved after repeating the hybridizations with whole sections from the original FFPE tumor block.
BCL2 breaks were detected in 52 of 384 (13.5%) DLBCL. BCL6 was rearranged in 116 of 404 (28.7%) samples, and 11 of 367 DLBCL harbored dual translocations involving BCL2 and BCL6 (3.0%). Rearrangements of MYC were found in 36 of 407 specimens (8.8%). Analyzing patients with measurements for each of the 3 breaks, the MYC rearrangement was the sole translocation in 14 of 35 (40.0%) tumor samples, while 21 cases (60.0%) harbored additional breaks in BCL2, BCL6, or both. Seventeen of 350 DLBCL (4.9%) had breaks of MYC and BCL2, and 8 of 350 (2.3%) of MYC and BCL6. A triple-hit constellation targeting BCL2, BCL6, and MYC simultaneously was detected in 4 of 350 samples (1.1%).
Immunophenotypes and cell of origin classification
DLBCL specimens with MYC translocations were more often CD10 positive and IRF4/MUM1 negative than were their MYC nonrearranged counterparts (84.4% vs 30.7% and 57.6% vs 14.7%, respectively) (P < .001 for both). A higher proportion of MYC break-positive tumors had a Ki67 index > 90% in comparison with MYC break-negative tumors (15 of 34 [44.1%] vs 68 of 305 [22.3%]; P = .005). For the cutpoint of 75% Ki67 positive cells investigated for EFS and OS, we did not observe a significant result regarding MYC-break (P = .468). BCL2 (32.4% vs 3.1%) and MYC breaks (18.9% vs 2.4%) were more frequent in GCB-DLBCL (P < .001 for both). In contrast, BCL6 breaks were more often observed in non-GCB DLBCL (34.7% vs 21.9%; P = .012). No significant differences in the distribution of breaks were observed between centroblastic and immunoblastic DLBCL or DLBCL NOS.
MYC, BCL2, and BCL6 immunohistochemistry
MYC stainings were reliably interpretable in 283 samples. A multicenter trial had been performed to ensure the quality of MYC staining evaluation, involving 7 diagnostic centers for hematopathology in Germany and Switzerland. Analyzing the number of positive cells in 6 B-NHL samples, we found the results to be highly concordant (κ-value of 0.69 for MYC overexpressed or not overexpressed tumors; data not shown). Ninety tumors (31.8%) showed overexpression (MYChigh) with samples harboring between 41% and 100% stained nuclei. The numbers of MYChigh cases were 18 of 26 (69.2%) in the MYC translocation-positive group, and 67 of 241 (27.8%) in the translocation-negative group (P < .001). Eight translocation-positive DLBCL, therefore, were MYClow. MYChigh samples harbored more tumors with a Ki67 index > 75% in comparison with MYClow tumors (73 of 83 [88.0%] vs 114 of 169 [67.5%], P < .001). No significant differences with respect to MYChigh protein expression were noted between GCB-DLBCL and non-GCB-DLBCL, and between centroblastic and immunoblastic DLBCL. Out of 442 patients with available FISH data, 313 of 393 (79.6%) were BCL2high (>0%), and 284 of 343 (82.8%) tumors were BCL6high (>25%). BCL2high was more frequent in non-GCB (154 of 177; 87.0%) than GCB-DLBCL (107 of 152; 70.4%) (P < .001), and BCL6high was observed in 130 of 177 non-GCB (73.4%) and in 137 of 143 (95.8%) GCB (P < .001). Out of 8 MYC break-positive patients with MYClow protein expression, 7 of 7 with IHC data were BCL2high and 6/7 were BCL6high. Within this group of patients, only 3 events occurred in EFS or OS.
Survival analysis
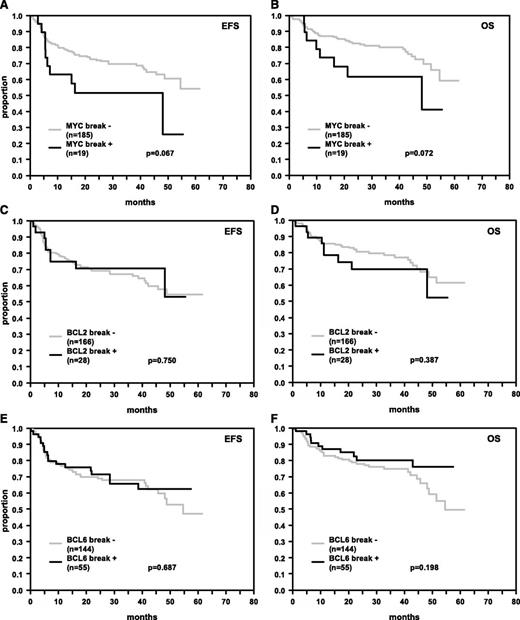
In the entire patient cohort, the presence of MYC translocations constituted a significant risk factor in univariate analysis (EFS: P = .025 [log rank], OS: P = .001 [log rank]) and in multivariate analyses adjusted for IPI factors (EFS: relative risk [RR] = 1.5 [95% CI: 0.9-2.4], P = .100, and OS: RR = 2.1 [95% CI: 1.3-3.5], P = .003). Inclusion of an interaction term (rituximab × MYC break) showed the difference in EFS/OS between MYC break-positive and -negative tumors within subgroups treated with or without rituximab to be in similar order (EFS: RR = 1.0; OS: RR = 0.7). Therefore, we observed relevant EFS and OS differences for patients with and without MYC translocation. Because of the small sample size especially of patients with MYC translocations in the rituximab-treated group, these differences did not reach statistical significance. The results for rituximab-treated patients are shown in Figure 1A-B.
EFS and OS in R-CHOP-treated DLBCL patients. Patients’ tumors were hybridized for breaks in MYC (A, B), BCL2 (C, D), and BCL6 (E, F). The numbers of patients showing breakage within these genes and P values are indicated.
EFS and OS in R-CHOP-treated DLBCL patients. Patients’ tumors were hybridized for breaks in MYC (A, B), BCL2 (C, D), and BCL6 (E, F). The numbers of patients showing breakage within these genes and P values are indicated.
In contrast to MYC translocations, breaks in BCL2 (Figure 1C-D) or BCL6 (Figure 1E-F) genes did not predict EFS and OS in univariate and multivariate analyses adjusted for the IPI factors in rituximab-treated patients (Table 2). We observed similar results for patients treated without rituximab (data not shown). In a multivariate Cox regression model for patients treated with rituximab including MYC, BCL2, and BCL6 breaks adjusted for IPI factors, neither BCL2 nor BCL6 breaks were independent prognostic factors for EFS (BCL2: RR = 0.8, P = .505; BCL6: RR = 1.0, P = .947) and OS (BCL2: RR = 1.3, P = .553; BCL6: RR = 0.7, P = .296). In addition, in our cohort, there were only a few double-hit lymphomas (with simultaneous breaks in MYC and BCL2 or BCL6 genes) among rituximab-treated patients, and therefore the confidence intervals for the 3-year OS and EFS rates were broad for these double-hit positive cases (MYC+/BCL2+ and MYC+/BCL6+) (EFS: 38.1% and 50.0%; 95% CI: 0.0-77.1 and 1.0-99.0; OS: 35.7% and 75.0%; 95% CI: 0.0-74.5 and 32.5-100.0; n = 7 and n = 4) in comparison with double-hit negative cases (MYC+/BCL2− and MYC+/BCL6−) (EFS: 58.3% and 53.3%; 95% CI: 30.5-86.1 and 28.0-78.6; OS: 75.0% and 59.3%; 95% CI: 50.5-99.5 and 34.0-84.6; n = 12 and n = 15). On the basis of the data obtained in the present patient cohort, therefore, no meaningful statement as to the risk impact of the double-hit feature could be made. In addition, Cox regression models including MYC break-only tumors, without additional breaks in BCL2 or BCL6, were unstable because of small sample size (data not shown).
Cox models adjusted for IPI factors for EFS and OS for patients treated with rituximab
| . | n* . | EFS . | OS . | |||||
|---|---|---|---|---|---|---|---|---|
| . | Positive . | Negative . | RR . | 95% CI . | P value . | RR . | 95% CI . | P value . |
| FISH | ||||||||
| MYC break + | 19 | 185 | 1.6 | 0.8-3.2 | 0.162 | 1.8 | .8-3.9 | 0.132 |
| BCL2 break + | 28 | 166 | 0.9 | 0.5-1.8 | 0.800 | 1.5 | 0.7-3.0 | 0.302 |
| BCL6 break + | 55 | 144 | 0.8 | 0.4-1.3 | 0.364 | 0.5 | 0.3-1.1 | 0.078 |
| IHC | ||||||||
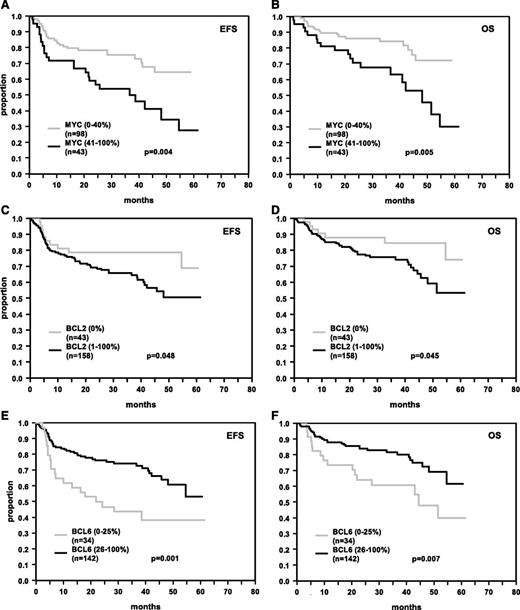
| MYC IHC (41%-100%) | 43 | 98 | 2.2 | 1.3-3.9 | 0.005 | 2.5 | 1.3-4.8 | 0.005 |
| BCL2 IHC (1%-100%) | 158 | 43 | 1.9 | 1.0-3.8 | 0.064 | 2.4 | 1.0-5.5 | 0.039 |
| BCL6 IHC (0%-25%) | 142 | 34 | 2.2 | 1.3-3.8 | 0.003 | 2.1 | 1.1-3.8 | 0.020 |
| IHC, model 1, n = 137 | ||||||||
| MYC IHC (41%-100%) | 43 | 94 | 2.2 | 1.2-3.8 | 0.007 | 2.4 | 1.2-4.6 | 0.013 |
| BCL2 IHC (1%-100%) | 110 | 27 | 3.4 | 1.3-9.0 | 0.013 | 5.1 | 1.4-18.0 | 0.012 |
| IHC, model 2, n= 138 | ||||||||
| MYC IHC (41%-100%) | 43 | 95 | 2.2 | 1.3-3.9 | 0.006 | 2.4 | 1.2-4.8 | 0.010 |
| BCL6 IHC (0%-25%) | 113 | 25 | 2.3 | 1.2-4.4 | 0.009 | 1.8 | 0.9-3.9 | 0.115 |
| IHC, model 3, n= 170 | ||||||||
| BCL2 IHC (1%-100%) | 132 | 38 | 1.5 | 0.8-3.1 | 0.228 | 1.9 | 0.8-4.5 | 0.144 |
| BCL6 IHC (0%-25%) | 136 | 34 | 2.2 | 1.3-3.8 | 0.004 | 2.0 | 1.1-3.7 | 0.035 |
| IHC, model 4, n = 135 | ||||||||
| MYC IHC (41%-100%) | 43 | 92 | 2.2 | 1.2-3.9 | 0.009 | 2.3 | 1.2-4.7 | 0.019 |
| BCL2 IHC (1%-100%) | 108 | 27 | 3.0 | 1.1-7.8 | 0.029 | 4.5 | 1.3-16.2 | 0.020 |
| BCL6 IHC (0%-25%) | 110 | 25 | 2.1 | 1.1-3.9 | 0.025 | 1.5 | 0.7-3.2 | 0.338 |
| FISH/IHC, model 1, n = 133 | ||||||||
| MYC break + | 14 | 119 | 1.5 | 0.6-3.4 | 0.361 | 2.1 | 0.8-5.5 | 0.151 |
| MYC IHC (41%-100%) | 41 | 92 | 2.0 | 1.1-3.7 | 0.030 | 2.1 | 1.0-4.4 | 0.063 |
| FISH/IHC, model 2, n = 186 | ||||||||
| MYC break + | 18 | 168 | 1.5 | 0.7-3.1 | 0.272 | 1.8 | 0.8-4.0 | 0.142 |
| BCL2 IHC (1%-100%) | 144 | 42 | 2.1 | 1.0-4.4 | 0.040 | 2.7 | 1.1-6.7 | 0.029 |
| FISH/IHC, model 3, n = 164 | ||||||||
| MYC break + | 19 | 145 | 1.9 | 0.9-3.8 | 0.074 | 2.1 | 1.0-4.7 | 0.062 |
| BCL6 IHC (0%-25%) | 131 | 33 | 2.4 | 1.4-4.1 | 0.003 | 2.4 | 1.2-4.5 | 0.009 |
| FISH/IHC, model 4, n = 158 | ||||||||
| MYC break + | 18 | 140 | 1.7 | 0.8-3.7 | 0.141 | 2.2 | 1.0-5.0 | 0.057 |
| BCL2 IHC (1%-100%) | 121 | 37 | 1.7 | 0.8-3.5 | 0.186 | 2.0 | 0.8-5.1 | 0.144 |
| BCL6 IHC (0%-25%) | 125 | 33 | 2.3 | 1.3–4.1 | 0.005 | 2.2 | 1.1–4.3 | 0.020 |
| FISH/IHC, model 5, n = 127 | ||||||||
| MYC break + | 14 | 113 | 1.5 | 0.6-3.5 | 0.402 | 1.8 | 0.6-5.3 | 0.259 |
| MYC IHC (41%-100%) | 41 | 86 | 1.9 | 1.0–3.7 | 0.052 | 1.8 | 0.8–4.2 | 0.159 |
| BCL2 IHC (1%-100%) | 100 | 27 | 2.7 | 1.0-7.2 | 0.049 | 3.8 | 1.0-14.1 | 0.045 |
| BCL6 IHC (0%-25%) | 102 | 25 | 2.2 | 1.2–4.3 | 0.016 | 1.8 | 0.8–4.0 | 0.156 |
| . | n* . | EFS . | OS . | |||||
|---|---|---|---|---|---|---|---|---|
| . | Positive . | Negative . | RR . | 95% CI . | P value . | RR . | 95% CI . | P value . |
| FISH | ||||||||
| MYC break + | 19 | 185 | 1.6 | 0.8-3.2 | 0.162 | 1.8 | .8-3.9 | 0.132 |
| BCL2 break + | 28 | 166 | 0.9 | 0.5-1.8 | 0.800 | 1.5 | 0.7-3.0 | 0.302 |
| BCL6 break + | 55 | 144 | 0.8 | 0.4-1.3 | 0.364 | 0.5 | 0.3-1.1 | 0.078 |
| IHC | ||||||||
| MYC IHC (41%-100%) | 43 | 98 | 2.2 | 1.3-3.9 | 0.005 | 2.5 | 1.3-4.8 | 0.005 |
| BCL2 IHC (1%-100%) | 158 | 43 | 1.9 | 1.0-3.8 | 0.064 | 2.4 | 1.0-5.5 | 0.039 |
| BCL6 IHC (0%-25%) | 142 | 34 | 2.2 | 1.3-3.8 | 0.003 | 2.1 | 1.1-3.8 | 0.020 |
| IHC, model 1, n = 137 | ||||||||
| MYC IHC (41%-100%) | 43 | 94 | 2.2 | 1.2-3.8 | 0.007 | 2.4 | 1.2-4.6 | 0.013 |
| BCL2 IHC (1%-100%) | 110 | 27 | 3.4 | 1.3-9.0 | 0.013 | 5.1 | 1.4-18.0 | 0.012 |
| IHC, model 2, n= 138 | ||||||||
| MYC IHC (41%-100%) | 43 | 95 | 2.2 | 1.3-3.9 | 0.006 | 2.4 | 1.2-4.8 | 0.010 |
| BCL6 IHC (0%-25%) | 113 | 25 | 2.3 | 1.2-4.4 | 0.009 | 1.8 | 0.9-3.9 | 0.115 |
| IHC, model 3, n= 170 | ||||||||
| BCL2 IHC (1%-100%) | 132 | 38 | 1.5 | 0.8-3.1 | 0.228 | 1.9 | 0.8-4.5 | 0.144 |
| BCL6 IHC (0%-25%) | 136 | 34 | 2.2 | 1.3-3.8 | 0.004 | 2.0 | 1.1-3.7 | 0.035 |
| IHC, model 4, n = 135 | ||||||||
| MYC IHC (41%-100%) | 43 | 92 | 2.2 | 1.2-3.9 | 0.009 | 2.3 | 1.2-4.7 | 0.019 |
| BCL2 IHC (1%-100%) | 108 | 27 | 3.0 | 1.1-7.8 | 0.029 | 4.5 | 1.3-16.2 | 0.020 |
| BCL6 IHC (0%-25%) | 110 | 25 | 2.1 | 1.1-3.9 | 0.025 | 1.5 | 0.7-3.2 | 0.338 |
| FISH/IHC, model 1, n = 133 | ||||||||
| MYC break + | 14 | 119 | 1.5 | 0.6-3.4 | 0.361 | 2.1 | 0.8-5.5 | 0.151 |
| MYC IHC (41%-100%) | 41 | 92 | 2.0 | 1.1-3.7 | 0.030 | 2.1 | 1.0-4.4 | 0.063 |
| FISH/IHC, model 2, n = 186 | ||||||||
| MYC break + | 18 | 168 | 1.5 | 0.7-3.1 | 0.272 | 1.8 | 0.8-4.0 | 0.142 |
| BCL2 IHC (1%-100%) | 144 | 42 | 2.1 | 1.0-4.4 | 0.040 | 2.7 | 1.1-6.7 | 0.029 |
| FISH/IHC, model 3, n = 164 | ||||||||
| MYC break + | 19 | 145 | 1.9 | 0.9-3.8 | 0.074 | 2.1 | 1.0-4.7 | 0.062 |
| BCL6 IHC (0%-25%) | 131 | 33 | 2.4 | 1.4-4.1 | 0.003 | 2.4 | 1.2-4.5 | 0.009 |
| FISH/IHC, model 4, n = 158 | ||||||||
| MYC break + | 18 | 140 | 1.7 | 0.8-3.7 | 0.141 | 2.2 | 1.0-5.0 | 0.057 |
| BCL2 IHC (1%-100%) | 121 | 37 | 1.7 | 0.8-3.5 | 0.186 | 2.0 | 0.8-5.1 | 0.144 |
| BCL6 IHC (0%-25%) | 125 | 33 | 2.3 | 1.3–4.1 | 0.005 | 2.2 | 1.1–4.3 | 0.020 |
| FISH/IHC, model 5, n = 127 | ||||||||
| MYC break + | 14 | 113 | 1.5 | 0.6-3.5 | 0.402 | 1.8 | 0.6-5.3 | 0.259 |
| MYC IHC (41%-100%) | 41 | 86 | 1.9 | 1.0–3.7 | 0.052 | 1.8 | 0.8–4.2 | 0.159 |
| BCL2 IHC (1%-100%) | 100 | 27 | 2.7 | 1.0-7.2 | 0.049 | 3.8 | 1.0-14.1 | 0.045 |
| BCL6 IHC (0%-25%) | 102 | 25 | 2.2 | 1.2–4.3 | 0.016 | 1.8 | 0.8–4.0 | 0.156 |
Sample sizes differ because of complete data set per Cox model.
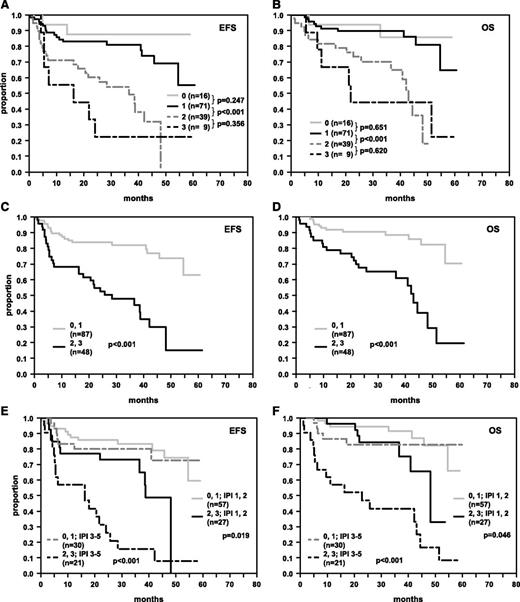
Protein overexpression of MYC (MYChigh) was an adverse prognostic factor for R-CHOP (EFS P = .004; OS P = .005; Figure 2A-B), but not for CHOP-treated patients (EFS P = .140; OS P = .941; data not shown). MYC overexpression retained its importance in R-CHOP-treated patients after adjusting for the IPI factors (EFS: RR = 2.2 [95% CI: 1.3-3.9], P = .005, and OS: RR = 2.5 [95% CI: 1.3-4.8], P = .005) and after adjusting for the IPI factors and the presence of MYC breaks (EFS: RR = 2.0 [95% CI: 1.1-3.7], P = .030, and OS: RR = 2.1 [95% CI: 1.0-4.4], P = .063) (Table 2). In accordance with our previously published results, expression patterns of BCL2 (Figure 2C-D) and BCL6 (Figure 2E-F) predicted EFS and OS in R-CHOP-treated patients within the RICOVER study.4 In Cox models adjusted for the IPI factors (Table 2), MYChigh, BCL2high, and BCL6low had prognostic relevance with relative risks greater than or equal to 1.5 across all models tested (IHC models 1-3). Most notably, inclusion of MYC, BCL2, and BCL6 expression in a multivariate Cox regression model adjusted for the IPI factors revealed all 3 factors as significant independent risk indicators in R-CHOP-treated patients (IHC model 4). Given that all 3 variables contributed in similar order in the multivariate Cox regression analyses, an IHC sum score of 0-3 was created, assigning an individual risk score of 0, 1, 2, or 3 for each patient according to the number of adverse features (MYChigh, BCL2high, BCL6low). Figure 3 illustrates the prognostic impact of the score for 4 groups (0, 1, 2, 3 adverse features), and for 2 groups (pooling the scores 0,1 and 2,3). Patients with a higher score had poorer survival. Within the IPI risk groups 1,2 and 3-5 the dichotomized IHC score separated patient groups with significant survival differences. Of pivotal importance, we identified a poor prognosis group (15% of patients) within the IPI risk group 3-5, with a 3-year EFS and OS of only 15.6% and 41.6%, respectively (Figure 3E-F). Cox regression analyses also including the morphological classifier (immunoblastic vs centroblastic) could not be undertaken because of the small sample size for patients with immunoblastic subtype within the Cox model (n = 9).
EFS and OS in R-CHOP-treated DLBCL patients. Patients’ tumors were stained for MYC (A, B), BCL2 (C, D), and BCL6 (E, F). The numbers of patients showing negative or positive stainings (MYC > 40%, BCL2 > 0%, BCL6 > 25%), cutoff values, and P values are indicated.
EFS and OS in R-CHOP-treated DLBCL patients. Patients’ tumors were stained for MYC (A, B), BCL2 (C, D), and BCL6 (E, F). The numbers of patients showing negative or positive stainings (MYC > 40%, BCL2 > 0%, BCL6 > 25%), cutoff values, and P values are indicated.
EFS and OS in R-CHOP-treated DLBCL patients whose tumors were stratified according to the number of adverse features in immunohistochemical stainings for MYChigh, BCL2high, and BCL6low. (A, B) Survival curves of patients stratified according to the presence of 0, 1, 2, or 3 adverse features. (C, D) Survival curves of patients who were stratified according to the presence of 0 or 1 and >1 adverse features. (E, F) Survival curves of patients stratified according to the presence of 0 or 1 and >1 adverse features and IPI groups 1,2 and 3-5. The numbers of patients within the respective groups and P values are indicated.
EFS and OS in R-CHOP-treated DLBCL patients whose tumors were stratified according to the number of adverse features in immunohistochemical stainings for MYChigh, BCL2high, and BCL6low. (A, B) Survival curves of patients stratified according to the presence of 0, 1, 2, or 3 adverse features. (C, D) Survival curves of patients who were stratified according to the presence of 0 or 1 and >1 adverse features. (E, F) Survival curves of patients stratified according to the presence of 0 or 1 and >1 adverse features and IPI groups 1,2 and 3-5. The numbers of patients within the respective groups and P values are indicated.
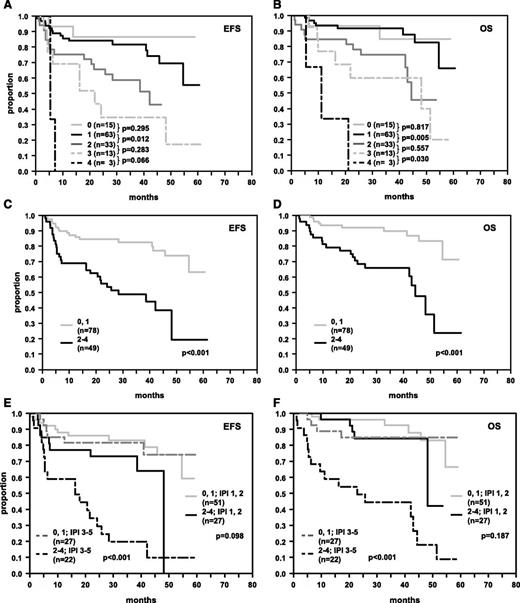
In view of our observation that not all tumors with detectable MYC breaks also showed MYChigh expression, we also tested Cox models including the variable “MYC breakage” (Table 2). In these models, the 3 immunohistochemical markers (MYChigh, BCL2high, and BCL6low) remained significant independent risk indicators. Relative risks in relevant order (EFS RR = 1.5, OS RR = 1.8) were also observed for MYC breaks (FISH/IHC model 5). In a combined algorithm including MYC, BCL2, and BCL6 protein expression patterns and MYC breakage (sum score ranging from 0 to 4 for each patient), patients with an index of >1 again had a significantly poorer EFS and OS. Figure 4 illustrates the prognostic impact of the FISH/IHC score for 5 groups (0, 1, 2, 3, 4; 4A,B), and in a dichotomized approach (groups 0,1 and 2,3,4; 4C,D), and within IPI groups (4E,F). The survival differences regarding EFS and OS as evident in the IHC or FISH/IHC score could also be observed within each of the IPI score groups 1,2,3,4-5, but suffered from small sample size within the single IPI groups (data not shown). The goodness of fit was comparable for both the IHC model and the FISH/IHC model (−2 log likelihood for EFS: 362.928 and 362.259). Chi-square tests were performed in order to test whether the cohort included either in the final IHC model or in the final FISH/IHC model is representative of the entire study population. No significant differences were observed when comparing relevant clinical characteristics of the IHC-score and the FISH/IHC-score patient samples to the entire RICOVER-60 cohort.
EFS and OS in R-CHOP-treated DLBCL patients whose tumors were stratified according to the number of adverse features in immunohistochemical stainings for MYChigh, BCL2high, and BCL6low and chromosomal translocations of MYC. (A, B) Survival curves of patients stratified according to the presence of 0, 1, 2, 3, or 4 adverse features. (C, D) Survival curves of patients who were stratified according to the presence of 0 or 1 and >1 adverse features. (E, F) Survival curves of patients stratified according to the presence of 0 or 1 and >1 adverse features and IPI groups 1,2 and 3-5. The numbers of patients within the respective groups and P values are indicated.
EFS and OS in R-CHOP-treated DLBCL patients whose tumors were stratified according to the number of adverse features in immunohistochemical stainings for MYChigh, BCL2high, and BCL6low and chromosomal translocations of MYC. (A, B) Survival curves of patients stratified according to the presence of 0, 1, 2, 3, or 4 adverse features. (C, D) Survival curves of patients who were stratified according to the presence of 0 or 1 and >1 adverse features. (E, F) Survival curves of patients stratified according to the presence of 0 or 1 and >1 adverse features and IPI groups 1,2 and 3-5. The numbers of patients within the respective groups and P values are indicated.
Discussion
The poor prognosis of CHOP-treated DLBCL patients whose tumors carry MYC rearrangements is well established.11,12,31 More recently, the negative prognostic impact of MYC translocations has also been confirmed in patients treated with R-CHOP. In these studies, MYC breaks have been encountered in roughly 10% of DLBCL.13,14,32 We have confirmed these data in the largest series analyzed to date comprising DLBCL samples from 442 patients treated in the prospective randomized RICOVER-60 study of the DSHNHL. Moreover, we are able to convincingly demonstrate that the analysis of translocations by FISH is feasible on TMAs in a highly reliable and reproducible manner. A total of 36 of 407 tumors (8.8%) had a MYC break, similar to the findings of Barrans and associates.13 Multivariate analysis disclosed MYC rearrangement as the sole significant genetic factor influencing prognosis in the RICOVER-60 trial, whereas breaks in BCL2 and BCL6 had no impact on survival.
MYC is a global transcription factor that has been reported to regulate up to 10% of genes within the human genome (reviewed by Slack and Gascoyne33 ). Deregulation of MYC has been described to produce an “avalanche” effect upon gene expression, thus further underlining its central role in cellular processes such as proliferation, differentiation, and metabolism.12,31 It is important to note, however, that deregulation of MYC is not solely based on translocations of the gene. The study of the German Molecular Mechanisms in Malignant Lymphoma Network described deregulated expression of MYC in a sizeable number of tumors with a Burkitt lymphoma expression profile lacking MYC translocation by FISH.12 Furthermore, these findings were also confirmed in recent studies showing significantly elevated MYC protein expression in tumors lacking translocations of the MYC gene.25,26,34 Interestingly enough, recent publications have suggested alternative mechanisms equally leading to upregulation of MYC. For example, microRNAs may regulate MYC expression, and accordingly, microRNA profiles have been found to be different between MYC-rearranged and non-rearranged Burkitt lymphomas.16,17 × Amplification of MYC has been shown to be associated with MYC overexpression and poorer prognosis.18,35 Taking together the above-mentioned findings, it is not surprising that we also encountered MYChigh protein expression in the obvious absence of detectable MYC rearrangements in a subset of DLBCL using a newly described monoclonal antibody.20,21,25,26,34
MYChigh protein expression was associated with inferior outcome both in the entire DLBCL cohort as well as in the R-CHOP-treated subgroup, in accordance with its central role in the regulation of thousands of genes in the genome, as also reported recently.25,26,34 Moreover, MYC protein expression retained its prognostic significance in R-CHOP-treated patients after adjusting for the IPI factors and—most notably—also after adjusting for the presence of MYC breaks. These data imply that MYC activation and deregulation represents a significant adverse prognostic factor in DLBCL, irrespective of the underlying cause. Interestingly, protein overexpression of MYC (MYChigh) was an adverse prognostic factor in R-CHOP-treated patients but not in CHOP-treated patients. The mechanism, by which MYC as well as BCL2 and BCL6 overexpression selectively influences the outcome of R-CHOP-treated patients in contrast to patients treated with CHOP only, is unknown. However, some possible links of rituximab and MYC have been described in the literature. Jazirehi and colleagues reported the rituximab-dependent inhibition of Raf-MEK1/2-ERK1/2 signaling, which sensitizes B-cells to rituximab treatment.36 The activation of MYC is functionally linked to the ERK1/2 pathway,37,38 and very interestingly, it has been shown that the inactivation of ERK1/2 signaling results in dephosphorylation and inhibition of MYC.39
We have previously shown that the use of the Hans classifier30 failed to identify patients at risk in the R-CHOP arm of the RICOVER study, while immunoblastic morphology had emerged as a robust negative prognostic factor.4 We here show that the integration of MYC protein expression and the MYC translocation status into existing survival predictors in DLBCL are valuable adjunct markers. In our analysis, MYChigh protein expression (or MYC rearrangement status) in concert with BCL2high and BCL6low status emerged as critical prognostic variables with comparable relative risks for both EFS and OS in DLBCL. A simple scoring approach that sums up combinations of these biological features was able to predict survival of DLBCL under R-CHOP treatment in addition to the well-established IPI factors. Specifically, patients with a sum score of >1 had an inferior survival both in the low-risk and high-risk IPI groups. Of note, the prognostic value of this combined IHC or IHC/FISH approach was particularly pronounced in the clinically adverse IPI 3-5 group, in which the 3-year EFS of the patient group identified at risk (IHC score 2,3 or IHC/FISH score 2-4) was 15% only. For this patient population, treatment strategies other than conventional R-CHOP are warranted. It is highly interesting to note that 2 recent studies also came to the conclusion that the dual deregulation of MYC and BCL2 evidenced by protein overexpression does potentiate the negative prognostic impact of the 2 factors, identifying cases of inferior OS and EFS when both proteins are overexpressed. Most notably, the study by Johnson and colleagues showed that this effect was also significant on the gene expression level, and in both studies a number of MYC protein expression positive cases without MYC translocation were identified.25,26 The importance of immunoblastic morphology could not be tested owing to the small sample size for patients with immunoblastic subtype within the Cox model (n = 9). In the RICOVER-60 study, inclusion of MYC break analysis via FISH to the MYC/BCL2/BCL6 protein expression model added little to its predictive value. As detailed above, the populations at risk identified with use of the IHC or the IHC/FISH classifier are greatly overlapping, and the goodness of fit was comparable for the IHC model and the FISH/IHC model (−2 log likelihood for EFS: 362.928 and 362.259). However, not all MYC translocated DLBCL samples showed a significant (>40%) MYC protein staining pattern. Paying tribute to the fact that only recent and as yet incomplete experience has been gained for immunohistochemical stainings with the anti-MYC antibody,20,21,25,26,34 at the moment we strongly advise the use of a combined MYC-FISH and IHC model. Nevertheless, further validation studies need to be performed to get insights into the applicability and practicability of MYC immunohistochemistry in the future. A possible compromise for the moment might be to use MYC staining as a screening tool first, and then hybridize all cases with MYC protein expression >20%. Taking this into account and admitting that FISH still represents a time-consuming and expensive method, and because the analysis of MYC protein expression by immunohistochemistry is easily performed in a routine clinical setting, its continued value as a predictive marker will have to be established. Nevertheless, screening for MYC deregulation via immunohistochemistry for MYC protein expression represents an attractive tool in order to identify patients at risk in everyday practice. On the other hand, the sensitivity of MYC staining in recognizing a breakpoint of MYC, according to our data, would be only high, if a threshold of 20% was chosen. Using this cutoff, FISH analysis for MYC could have been “safely” omitted in 90 of 241 (37.3%) patients in our study with a sensitivity of 92.3%. It has to be kept in mind, however, that immunohistochemistry, especially regarding the assessment of BCL6 expression, has recently been shown to be difficult to reproduce, arguing for the addition of MYC FISH to the model, resulting in a more robust prognostic score.40
Generally, our results had been generated in patients ages 61-80 years. Given the lack of comparable investigations reported to date in younger as well as in older patients beyond 80 years, it remains to be clarified whether the prognostic implications of chromosomal aberrations and protein expression patterns reported here will apply to these age groups as well.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors would like to thank Daniela Rauh (Stuttgart), Beate Mann (IMISE Leipzig), Inge Klier (Würzburg), Olivera Batic and Christiane Stange (Kiel), Michaela Buck (Ulm), Erika Berg (Berlin), Katharina Vogel and Ilona Schliephake (Lübeck), and Ralf Lieberz (Frankfurt) for expert technical assistance.
This work was supported by the Deutsche Krebshilfe and by the Robert Bosch Stiftung, Stuttgart, Germany. S.C. was supported by the Swiss Group for Clinical Research (SAKK).
Authorship
Contribution: H.H., M.Z., M.L., A.R., and G.O. conceived and designed the experiment. A.R., W.K., A.C.F., H.S., M.L.H., S.C., P.M., and G.O. were on the pathology reference panel. G.O., W.K., A.C.F., S.C., H.S., M.L.H., P.M., T.F.E.B., H.W.B., M.P., N.S., L.T., and A.R. provided study materials or patients. H.H., M.Z., C.B., C.S., W.K., H.W.B., A.C.F., H.S., M.H., M.L.H., T.F.E.B., P.M., M.P., N.S., L.T., R.S., S.C., A.R., and G.O. collected and assembled the data. M.Z. and M.L. performed the biometrics. H.H., M.Z., M.L., A.R., and G.O. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof. Dr. German Ott, Department of Clinical Pathology, Robert-Bosch-Krankenhaus, Auerbachstrasse 110, 70376 Stuttgart, Germany; e-mail: german.ott@rbk.de.
References
Author notes
H.H. and M.Z. contributed equally to this work.
A.R. and G.O. are joint senior authors.