Key Points
This research is the first to document that human eosinophils undergo extracellular DNA trap cell death.
This research revealed a process by which eosinophils undergo cytolysis to liberate intact cell-free and secretion-competent granules.
Abstract
Eosinophils release their granule proteins extracellularly through exocytosis, piecemeal degranulation, or cytolytic degranulation. Findings in diverse human eosinophilic diseases of intact extracellular eosinophil granules, either free or clustered, indicate that eosinophil cytolysis occurs in vivo, but the mechanisms and consequences of lytic eosinophil degranulation are poorly understood. We demonstrate that activated human eosinophils can undergo extracellular DNA trap cell death (ETosis) that cytolytically releases free eosinophil granules. Eosinophil ETosis (EETosis), in response to immobilized immunoglobulins (IgG, IgA), cytokines with platelet activating factor, calcium ionophore, or phorbol myristate acetate, develops within 120 minutes in a reduced NADP (NADPH) oxidase-dependent manner. Initially, nuclear lobular formation is lost and some granules are released by budding off from the cell as plasma membrane–enveloped clusters. Following nuclear chromatolysis, plasma membrane lysis liberates DNA that forms weblike extracellular DNA nets and releases free intact granules. EETosis-released eosinophil granules, still retaining eosinophil cationic granule proteins, can be activated to secrete when stimulated with CC chemokine ligand 11 (eotaxin-1). Our results indicate that an active NADPH oxidase-dependent mechanism of cytolytic, nonapoptotic eosinophil death initiates nuclear chromatolysis that eventuates in the release of intact secretion-competent granules and the formation of extracellular DNA nets.
Introduction
Human eosinophils, leukocytes notably associated with allergic, anthelmintic parasite, and other immune responses,1-3 contain an abundant singular population of crystalloid-bearing granules. As intracellular organelles, these granules are central to the functional responses of eosinophils in that eosinophil granules house preformed protein stores of (1) 4 major cationic proteins, including eosinophil cationic protein (ECP), major basic protein (MBP), and eosinophil peroxidase (EPO); (2) hydrolytic enzymes; and (3) over 4 dozen cytokines, chemokines, and growth factors.4,5 Intact eosinophils may secrete their granule proteins by classic exocytosis (principally on the surfaces of large, multicellular helminths) or more commonly by piecemeal degranulation.4
Unlike granules from other granule-containing leukocytes, eosinophil granules have long been recognized to be present extracellularly in tissues and sputum associated with diverse eosinophil-associated diseases. Extracellular eosinophil granules have been detected by immunostaining for their cationic proteins and/or by ultrastructural studies that demonstrated intact granules still bound by their granule-delimiting membranes. Free extracellular eosinophil granules have been documented in diverse diseases, including atopic dermatitis and nasal allergy, and they have been correlated with the severity of urticaria.4,6 Since the late 1800s, free eosinophil granules have been noted in the sputum of asthmatics, and clusters of free eosinophil granules (termed “Cfegs”) have been documented in human asthma and experimental guinea pig models of asthma.7
The release of intact, membrane-bound granules occurs via an enigmatic mode of eosinophil “degranulation” that arises from the cytolysis of eosinophils. Prior in vivo studies revealed that (1) lytic eosinophils, noted ultrastructurally by chromatolysis and loss of plasma membrane integrity, were frequently observed in human airway specimens rather than apoptotic cells,8,9 (2) the numbers of free granules increased severalfold within an hour after allergen-induced airway provocation,7 and (3) released granules exhibited little evidence of loss of their granule contents.10 Collectively, the many in vivo findings of free extracellular eosinophil granules suggested that a process exists for human eosinophils to undergo nonapoptotic but cytolytic cell death that liberated intact extracellular granules.
Defining the means by which eosinophils may release extracellularly intact granules is more cogent with our recent findings, in which isolated eosinophil granules remain secretion competent. Cell-free eosinophil granules express ligand-binding cytokine, chemokine, and eicosanoid receptors on the surface of their delimiting membranes.11,12 Cell-free human and mouse eosinophil granules with receptor-mediated activation of intragranule signaling pathways can directly secrete selected granule-derived proteins, including ECP, EPO, ribonucleases, and cytokines, for example, the interleukins IL-4 and IL-6.12-14 Thus, the local tissue release of cell-free, secretion-competent eosinophil granules, secondary to eosinophil lysis, might constitute a means by which postmortem eosinophils could continue to provide immunoregulatory, pro-inflammatory, and other immunopathogenic stimuli. Despite these observations, the mechanisms of the eosinophil cytolytic release of their intact granules are not well known.
Recently, an active form of cell death, namely, extracellular DNA trap cell death (called ETosis15 ), has been recognized in neutrophils16 and mast cells.17 In these cells, ETosis develops with time, often over 1 or more hours, and is morphologically distinct from other classic cell death processes, including apoptosis and necrosis.18 In contrast to apoptosis, nuclear condensation and DNA fragmentation do not occur. Instead, nuclear chromatin decondenses in the cytoplasm.19 Finally, rupture of the plasma membrane releases nuclear DNA to form extracellular DNA nets, which for neutrophils bind free antimicrobial molecules such as granule proteins and histones.20 Reduced NADP (NADPH)-oxidase-mediated production of reactive oxygen species (ROS) has an essential role in the activation of ETosis.15 To date, while it has been shown that IL-5– or interferon-γ−primed human eosinophils, in response to certain agonists, can very rapidly and noncytolytically release mitochondrial, and not nuclear, DNA to form extracellular DNA nets,21 eosinophil ETosis has not been recognized.
Prior studies reported that the immobilized immunoglobulins (IgG, IgA) and the calcium ionophore A23187 induced eosinophil cell death morphologically, similar to lytic degranulation.22,23 In the present study, we evaluated whether eosinophil cytolysis is mediated by ETosis. We have now shown that activated human eosinophils undergo NADPH-oxidase-dependent ETosis and that during ETosis-mediated cytolysis, eosinophil granules are liberated both when contained within plasma membrane–bound clusters and as cell-free granules. Cell-free eosinophil granules retained their cationic proteins, and some were functionally competent to secrete in response to the chemokine ligand CCL11 (also known as eotaxtin-1).
Material and methods
Sample collection and cell preparations
Eosinophils were purified from 320 mL of blood from mildly atopic and healthy donors by negative selection, as described in Neves et al.14 In accordance with the Declaration of Helsinki, informed consent was obtained under Institutional Review Board–approved protocols. Briefly, after blood was collected into a 6% dextran saline solution (Pharmacosmos, Holbaek, Denmark) and red blood cells (RBCs) were sedimented, the upper leukocyte-containing region was collected and centrifuged over Ficoll-Paque (GE Healthcare, Pittsburgh, PA). Eosinophils were isolated from granulocyte pellets by incubation with a depletion antibody (Ab) cocktail (StemSep, Stemcell Technologies, Vancouver, BC, Canada) followed by passage over magnetized columns (Miltenyi Biotec, Auburn, CA). The purity of isolated eosinophils was >97% of nucleated cells with ≤3% contaminating RBCs, and eosinophil viability was >99%, as determined by microscopic analyses. Separately, RBCs (>99% purity) were purified from the dextran sedimented fraction, as described in Aoshiba et al.24 Tissue biopsy samples were obtained during clinical evaluations from the left frontal sinus and the skin from a patient with allergic sinusitis and a patient with hypereosinophilic syndrome (negative for FIP1-like 1/platelet-derived growth factor-α mutation), respectively.
Detection of extracellular chromatin structure and cell death
Purified eosinophils were stimulated with A23187 (Sigma-Aldrich, St. Louis, MO), PMA (phorbol 12-myristate 13-acetate, Sigma-Aldrich), IL-5 (R&D Systems, Minneapolis, MN), GM-CSF (R&D Systems), and/or platelet activating factor (PAF; Enzo Life Sciences, Farmingdale, NY) in poly-L-lysine–coated chambered cover glasses (Nalge Nunc, Rochester, NY) or 96-well, flat-bottomed tissue culture plates in 0.1% bovine serum albumin (BSA, wt/vol; Sigma-Aldrich) or as stated in fetal bovine serum (FBS; Gibco, Grand Island, NY) containing phenol red-free HEPES-buffered RPMI 1640 medium. Human IgG or IgA (Sigma-Aldrich), diluted with phosphate-buffered saline (Gibco) in indicated concentrations, was coated on culture plates for 3 hours at 37°C. For detection of extracellular chromatin structure, eosinophils (2 × 105 cells per well) were seeded on chamber slides and stimulated with the indicated conditions followed by fixation with 3% paraformaldehyde (PFO, Electron MIcroscopy Sciences, Hatfield, PA) without permeabilization. Eosinophils were incubated with mouse Abs against histones (recognizing histones H1, H2A, H2B, and H4, 1:500, 45 minutes at room temperature (RT); United States Biological, Swampscott, MA) followed by goat antimouse IgG (1:100, 30 minutes at RT; Invitrogen, Carlsbad, CA) and propidium iodide (PI; Invitrogen). For quantification of cell death, CYTOX green (Invitrogen) was added to the medium, and CYTOX intensity was measured using the GloMax-Multi detection system (Promega, Madison, WI). The increase of CYTOX intensity was concurrent with cell death (confirmed using lactate dehydrogenase release). In some experiments, diphenyleneiodonium chloride (DPI, 20 μM; Sigma-Aldrich) was added to the culture medium.
Scanning electron microscopy (SEM)
Eosinophils, added to round coverslips coated with poly-L-lysine in culture plates, were stimulated with A23187 for 1 hour. Cells were fixed (1% PFO, 1.25% glutaraldehyde in 1 M sodium cacodylate buffer, pH 7.4) for 30 minutes at RT. Coverslip-adherent cells were postfixed with 1% osmium tetroxide in water for 1 hour and dehydrated in an ascending ethanol series from 50% (vol/vol) to absolute ethanol (10 minutes per step). Cells were then critical point dried in carbon dioxide. Coverslips were mounted on aluminum holders, sputtered with 5 nm gold, and analyzed in a scanning electron microscope (Quanta 200 FEG; FEI, Eindhoven, The Netherlands).
Immunostaining for eosinophil granule MBP
Eosinophils, seeded in 8-well Lab-Tek II chamber slides (Nalge Nunc) or immunoglobulin-coated Millicell EZ slides (Millipore), were stimulated for indicated time points and then fixed with 3% PFO for 10 minutes. For MBP staining, cells were permeabilized with 0.1% saponin for 10 minutes, incubated with primary mouse antihuman MBP Ab (5 μg/mL, clone AHE-2, 60 minutes at 4°C; BD Pharmingen), and followed by Alexa Fluor 488 conjugated Ab (goat antimouse IgG, 1:300, 30 minutes at RT; Invitrogen). Control Abs and PI were used for each experiment.
Τransmission electron microscopy (TEM)
Blood eosinophils were added to chamber slides, stimulated with 2 μM of A23187 for 1 hour, and immediately fixed in a mixture of freshly prepared aldehydes (1% PFO, 1.25% glutaraldehyde) in 1 M sodium cacodylate buffer, pH 7.4, for 30 minutes at RT. To obtain optimal morphology, cells directly on the slide surface were postfixed in 1% osmium tetroxide and processed as described in Melo et al.25,26 Biopsy samples were fixed for 4 hours at RT using the same fixative and processed for electron microscope as before.25,26 Sections were mounted on uncoated 200-mesh copper grids (Ted Pella, Redding, CA) before staining with lead citrate, and then viewed with a transmission electron microscope (CM10; Philips, Eindhoven, The Netherlands) at 60 kV.
Time-lapse microscopy
Eosinophils in chambered cover glasses were stimulated with immobilized IgG (1 mg/mL) for 40 minutes or 2 μM A23187 for 20 minutes in a carbon dioxide incubator. Chambered cover glasses were sealed with 37°C mineral oil before microscopic imaging, and morphological changes and granule release were evaluated using an inverted microscope (Eclipse TE300; Nikon, Tokyo, Japan) equipped with a digital camera in conjunction with iVision 4.0 image analysis software (BioVision, Exton, PA). The stage was preheated at 37°C, and phase-contrast images were recorded for 30 minutes (every 3 seconds) with a 100× objective. Images were sequenced into movies (every 0.03 seconds per image).
Evaluation of released eosinophil subcellular structures by using flow cytometry
Through the use of a modified fluorescent bead-count–based assay for microparticles,27 subcellular structures in eosinophil culture medium were counted and studied using FACScan (BD Biosciences, San Jose, CA). In some experiments, subcellular structures from eosinophils loaded with acridine orange (AO, 3 μM, 15 minutes; Sigma-Aldrich) and structures stained with annexin V (Invitrogen) were examined. Data were analyzed by Flowjo (version 9.03; Tree Star, Ashland, OR). Details are available in supplemental “Materials” (supplemental Figure 1).
Staining of granule-rich subcellular structures
After eosinophils were stimulated with 2 μM A23187 for 1 hour, culture medium was gently collected. Contaminating cells and DNA were removed by centrifugation at 200 × g for 10 minutes, followed by incubation for 10 minutes with DNase I (20 U/mL; New England Biolabs, Ipswich, MA). Thereafter, supernatants were centrifuged at 2500 × g for 10 minutes, and pelleted subcellular structures were stained as previously described in Neves et al.11 The gating strategy for flow cytometry was the same as for supplemental Figure 1. CellMask, LysoTracker, and CellTracker (Invitrogen) were used for microscopic analyses as described in supplemental “Materials” (supplemental Figure 7).
ECP measurements
ECP levels in supernatants and extraction buffer14 (Biosource, Camarillo, CA)-disrupted subcellular structures were quantified by ECP enzyme-linked immunosorbent assay kits (Medical and Biological Labs, Nagoya, Japan). Samples were frozen at −20°C until measurement.
Stimulation of subcellular granule-associated structures with CCL11
Eosinophils (5 to 7 × 106 cells) were stimulated with 2 μM A23187 for 1 hour in 0.1% BSA containing RPMI medium, followed by the addition of 20 U/mL DNase I for 30 minutes. Culture medium was collected, residual intact eosinophils were removed by centrifugation at 200 × g for 10 minutes, and granule-rich subcellular structures were pelleted by 2500 × g centrifugation for 10 minutes. Pelleted structures were washed and resuspended in 0.1% BSA RPMI medium. The absence of live, intact eosinophil contamination was confirmed by trypan blue staining. Released granule-associated structures were incubated with the indicated concentrations of CCL11 (R&D Systems) or 2 μM A23187 for 1 hour at 37°C.
Activation of granule-associated subcellular structures derived from AO-loaded eosinophils
After AO loading, eosinophils were stimulated for 1 hour with 2 μM A23187, and ETosis-released granule-rich subcellular structures were isolated by centrifugation. Cell-free AO-stained granules were stimulated as described previously in Neves et al with some modifications.14 After recovered structures were washed and resuspended in 0.1% BSA RPMI medium, they were spread on poly-L-lysine–coated slides and coverslipped. Stimuli (800 ng/mL CCL11 or 4 μM A23187) or control medium were introduced in 2-μl drops at the edge of the coverslips, enabling stimuli to diffuse under the coverslip. The responses of AO-stained granules were monitored by fluorescence microscopy using the fluorescein isothiocyanate band fluorescence filter. To prevent AO quenching during time-lapse microscopy, 3 neutral-density filters were used during data acquisition. Fluorescence images were recorded for 100 seconds (100 frames total) and acquired using a BX62 microscope (Olympus).
Statistical analysis
Results were expressed as mean ± SD. The differences of groups were evaluated using 2-tailed Student t test, with the level of statistical significance taken as P < .05.
Results
Human tissue eosinophils in vivo exhibit cytolysis accompanied by nuclear disruption and extracellular granule release
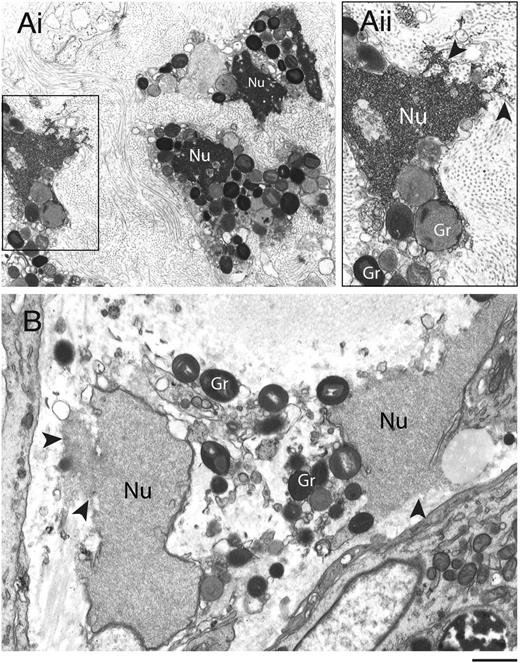
Disruptions of nuclear and plasma membranes (PMs) in cytolytic eosinophils have been observed in varied allergic and parasitic diseases.28,29 To ascertain the ultrastructural characteristics of eosinophil cytolysis in vivo, tissue-infiltrated eosinophils were evaluated using TEM (Figure 1). Eosinophils within biopsies from the sinus and the skin (from patients with allergic sinusitis and hypereosinophilic syndrome, respectively) showed similar characteristics of cytolytic degranulation with disrupted PMs and nuclear envelope membranes. In contrast to the nuclear chromatin condensation observed in apoptotic eosinophils,30 both distinct nuclear heterochromatin decondensation and release of nuclear contents into the extracellular space (Figure 1, arrowheads) were observed. Moreover, eosinophil granules released in vivo were intact and exhibited little evidence of loss of their granule contents (Figure 1).
Transmission electron micrographs of human tissue eosinophils from (A) allergic and (B) hypereosinophilic patients. Biopsy samples from the left frontal sinus and the skin were obtained from a patient with allergic sinusitis and a patient with hypereosinophilic syndrome (negative for FIP1-like 1/platelet-derived growth factor-α mutation), respectively. In (Ai), a biopsy of the frontal sinus shows infiltrated eosinophils. Note the disrupting nuclei (Nu) and extracellular free secretory granules. (Aii) is the boxed area of (Ai) seen at higher magnification. In (B), both free granules (Gr) and disrupted nuclei (Nu) are also observed in eosinophils from a skin biopsy. Arrowheads in (Aii) and (B) indicate releasing decondensed chromatin. Samples were fixed in a mixture of glutaraldehyde and PFO and prepared for conventional TEM as in “Materials and methods.” Scale bars represent 1.2µm (Ai), 700 nm (Aii), 600 nm (B).
Transmission electron micrographs of human tissue eosinophils from (A) allergic and (B) hypereosinophilic patients. Biopsy samples from the left frontal sinus and the skin were obtained from a patient with allergic sinusitis and a patient with hypereosinophilic syndrome (negative for FIP1-like 1/platelet-derived growth factor-α mutation), respectively. In (Ai), a biopsy of the frontal sinus shows infiltrated eosinophils. Note the disrupting nuclei (Nu) and extracellular free secretory granules. (Aii) is the boxed area of (Ai) seen at higher magnification. In (B), both free granules (Gr) and disrupted nuclei (Nu) are also observed in eosinophils from a skin biopsy. Arrowheads in (Aii) and (B) indicate releasing decondensed chromatin. Samples were fixed in a mixture of glutaraldehyde and PFO and prepared for conventional TEM as in “Materials and methods.” Scale bars represent 1.2µm (Ai), 700 nm (Aii), 600 nm (B).
Eosinophil ETosis (EETosis) releases nuclear DNA in response to various stimuli
In neutrophils, ETosis releases nuclear contents as filamentous chromatin structures, and neither apoptosis nor necrosis induces DNA nets.19 To test the hypothesis that immobilized IgG or IgA23 and A23187,22 previously shown to elicit eosinophil cytolysis, elicit ETosis, eosinophils were stained with antibodies against histones and fixed-cell permeable PI. Using nonpermeabilized cells, we were able to distinguish intact cells and extracellular histones (supplemental Figure 2). Immobilized IgG and IgA, like A23187, elicited the release of histone-positive DNA from disrupted cells (Figure 2A). In addition, PAF, in combination with IL-5 or GM-CSF, induced EETosis (Figure 2A, supplemental Figure 2A). PAF itself was a less potent and somewhat donor-dependent EETosis inducer, and IL-5 or GM-CSF alone did not induce EETosis (data not shown). PMA, an ETosis inducer for neutrophils,16 also induced EETosis.
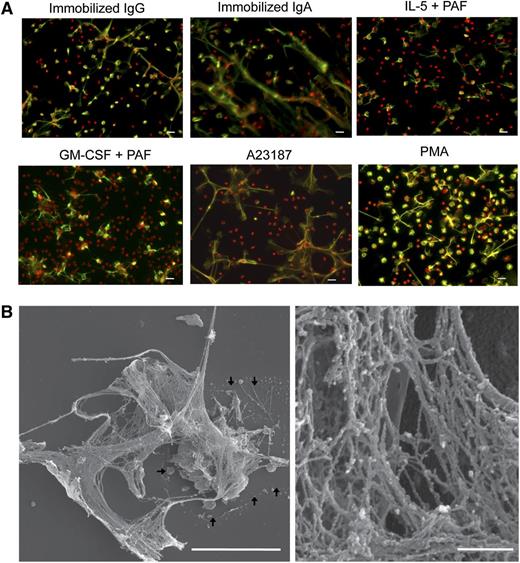
EETosis releases weblike chromatin structures from lytic cells. (A) Eosinophils were stimulated with the indicated stimuli in 0.1% BSA medium for 120 minutes or in A23187 for 60 minutes and fixed. EETosis was detected using extracellular histone staining without permeabilization, as described in “Materials and methods” and supplemental Figure 3. Green indicates histones, and red indicates DNA. Images were obtained using a BX62 microscope (20×, equipped with a Qimaging Rolera EM-C2 cooled digital camera (Surrey, BC, Canada) in conjunction with SlideBook 5.0 image analysis software (Intelligent Imaging Innovations, Denver, CO), or equipped with a Qimaging Retiga EXi cooled digital camera in conjunction with iVision image analysis software). Experiments were repeated with eosinophils from 3 independent donors with similar results. (B) Eosinophils were stimulated with 2 μM A23187 for 60 minutes and processed for SEM. Large DNA nets were released from lytic cells and often were associated with originating cells (outlined by arrows, left). Weblike DNA nets consisted of 25- to 35-nm diameter fibers aggregated into larger fibers (right). Scale bars represent 20 μm (A), 10 μm (B, left panel), and 500 nm (B, right panel).
EETosis releases weblike chromatin structures from lytic cells. (A) Eosinophils were stimulated with the indicated stimuli in 0.1% BSA medium for 120 minutes or in A23187 for 60 minutes and fixed. EETosis was detected using extracellular histone staining without permeabilization, as described in “Materials and methods” and supplemental Figure 3. Green indicates histones, and red indicates DNA. Images were obtained using a BX62 microscope (20×, equipped with a Qimaging Rolera EM-C2 cooled digital camera (Surrey, BC, Canada) in conjunction with SlideBook 5.0 image analysis software (Intelligent Imaging Innovations, Denver, CO), or equipped with a Qimaging Retiga EXi cooled digital camera in conjunction with iVision image analysis software). Experiments were repeated with eosinophils from 3 independent donors with similar results. (B) Eosinophils were stimulated with 2 μM A23187 for 60 minutes and processed for SEM. Large DNA nets were released from lytic cells and often were associated with originating cells (outlined by arrows, left). Weblike DNA nets consisted of 25- to 35-nm diameter fibers aggregated into larger fibers (right). Scale bars represent 20 μm (A), 10 μm (B, left panel), and 500 nm (B, right panel).
To ascertain differences among apoptosis, necrosis, and EETosis, we identified conditions that led to ∼20% dead cells (supplemental Figure 3A) mediated by necrosis, apoptosis, or EETosis by inducing cell death by brief heating, anti-Fas Abs,31 or A23187, respectively. Morphological changes were evaluated using phase-contrast microscopy and concomitant staining for annexin V and PI (supplemental Figure 3B). Anti-Fas–activated cells showed typical apoptotic morphology (cytoplasmic and nuclear condensation, budding) with bright annexin V–positive membranes and abundant released apoptotic bodies. Heated cells showed bleb formation, characteristic of necrotic cells.32 EETosis exhibited completely different appearances, releasing filamentous DNA and cell-free granules, with only weak staining for annexin V relative to Fas-activated cells. In heated or EETosis cells, surface annexin V single-positive cells were rarely observed, indicating that the phosphatidylserine (PS) redistribution typical during apoptosis was lacking. Of note, extracellular filamentous DNA was never observed in heated or Fas-activated cells. As assessed by the release of lactate dehydrogenase and histone staining, A23187-stimulated eosinophils exhibited cytolytic EETosis in an A23187 concentration-dependent manner (supplemental Figure 3C).
To study the process of nuclear shape changes during EETosis, we loaded live eosinophils with Hoechst 33342 dye, which stained nuclear DNA; cells were fixed at different time points. As shown in supplemental Figure 4, supplemental Video 1, and supplemental Video 2, initially the bilobular form of the eosinophil nucleus started to lose its shape, and thereafter the nuclear envelope disintegrated and DNA filled the cytoplasm. Finally, the PM ruptured, allowing the extrusion of the DNA net (confirmed by DNase treatment; data not shown). This process of nuclear shape change and dissolution was entirely consistent with neutrophil ETosis.19 A23187-induced EETosis developed more rapidly when compared with immobilized IgG and PMA (supplemental Figure 4). High-resolution SEM showed that DNA nets were associated with lytic cells (Figure 2B, left, arrows). DNA nets formed weblike structures that consisted mostly of fibers with diameters of 25 to 35 nm in conjunction with larger fibers (Figure 2B, right). These results indicate that activated eosinophil cell death is EETosis, which is distinct from necrosis and apoptosis.
EETosis is NADPH oxidase-dependent cell death
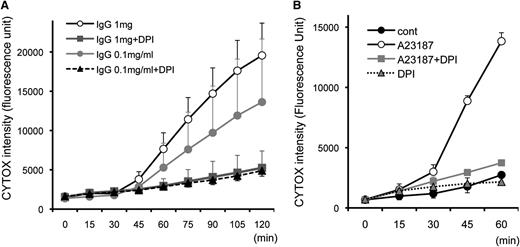
Neutrophil ETosis is dependent on ROS and inhibited by the NADPH oxidase inhibitor DPI.18 To monitor the kinetics of cell death and the effect of DPI, eosinophils were stimulated with immobilized IgG and A23187, and cell death was measured every 15 minutes (Figure 3). CYTOX intensity progressively increased 45 minutes after immobilized IgG stimulation in a concentration-dependent manner, and DPI completely abolished the effect of IgG- and A23187-stimulated eosinophils to undergo DPI-inhibitable cell death. Similar inhibition was observed with DPI when stimulated with other EETosis-inducing stimuli we tested (data not shown). In line with the time course of nuclear shape change (supplemental Figure 4), A23187-induced EETosis developed ∼30 minutes earlier when compared with IgG-induced EETosis (Figure 3B). As reported for ETosis in mast cells and neutrophils,17,19 both fetal bovine serum and BSA had concentration-dependent protective effects on A23187-induced EETosis (supplemental Figure 5A), likely based on their capacities to scavenge ROS. To further evaluate whether the generation of extracellular ROS was critical for EETosis, eosinophils were stimulated in the presence of the cell-impermeable ROS scavenging enzyme, catalase. Catalase inhibited A23187-induced eosinophil cytolysis and death (supplemental Figure 5B). RBCs, also known to scavenge ROS,24 likewise inhibited A23187-induced eosinophil cytolysis (supplemental Figure 5C).
EETosis developing within 120 minutes in an NADPH oxidase-dependent manner. Eosinophils (in 0.1% BSA and CYTOX containing RPMI medium) incubated with (A) 1 mg/mL immobilized IgG and (B) 2 μM A23187 with the indicated stimuli concentrations induced temporal EETosis cytolysis (extracellular CYTOX intensity) attributable to DPI-inhibitable NADPH oxidase activity. For (A), data are means ± SD, from 3 different donors, and for (B), data are means ± SD from 4 different donors.
EETosis developing within 120 minutes in an NADPH oxidase-dependent manner. Eosinophils (in 0.1% BSA and CYTOX containing RPMI medium) incubated with (A) 1 mg/mL immobilized IgG and (B) 2 μM A23187 with the indicated stimuli concentrations induced temporal EETosis cytolysis (extracellular CYTOX intensity) attributable to DPI-inhibitable NADPH oxidase activity. For (A), data are means ± SD, from 3 different donors, and for (B), data are means ± SD from 4 different donors.
EETosis releases intact granules and DNA nets predominantly free of granule proteins
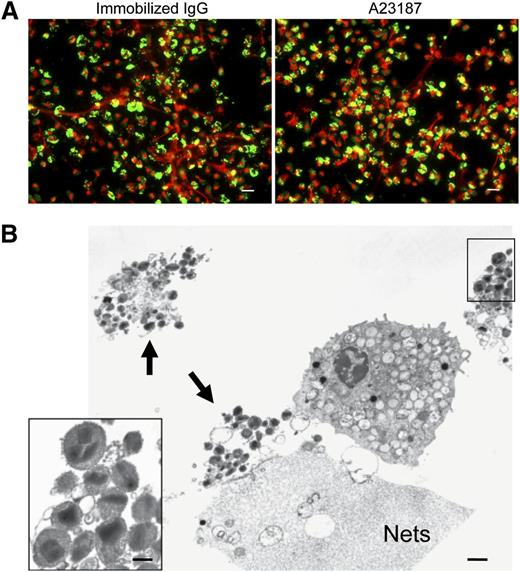
Neutrophils, diverse granule-derived proteins, many with antimicrobial activities, have been recognized to be bound as free proteins to extracellular DNA nets.16,33 Likewise, for the elicited release of mitochondrial DNA from eosinophils, eosinophil granule proteins have been found bound to extracellular DNA nets.21 To assess whether specific eosinophil granule proteins were released free or were otherwise bound to eosinophil DNA nets, EETosis cells were immunostained for MBP (Figure 4A). Notably, many regions of extracellular DNA nets lacked linear immunostaining for free MBP, and immunostaining was localized to smaller punctate foci or cell-associated domains, suggestive that eosinophil granule proteins might still be within granules. Immunostaining for EPO showed similar results (data not shown). Indeed, TEM analyses of EETosis revealed the presence of clusters of intact eosinophil granules (Figure 4B, arrowheads), and some were associated with DNA nets. In residual intact eosinophils, granules exhibited various degrees of electron density, likely indicating secretion of some of their contents. Ultrastructurally, cell-free eosinophil granules released from cytolytic cells retained their typical morphologies with intact granule-delimiting membranes and intact crystalloid cores and granule matrices (Figure 4B, insert).
Intact granules and limited granule protein localization at extracellular DNA nets released through EETosis-mediated cell lysis. (A) Eosinophil EETosis was induced using 1 mg/mL immobilized IgG (120 minutes) or 2 μM A23187 (60 minutes) and fixed and permeabilized, then stained for MBP (green) and DNA (PI, red). Most released DNA nets did not contain free granule MBP protein and instead showed punctate or cell-associated staining. Images were obtained with a BX62 Olympus upright microscope, 20× UPlanApo objective with a numerical aperture of 0.70, coupled to a Hamamatsu Orca-AG fire-wire cooled digital camera (Hamamatsu Photonics, Hamamatsu, Japan; images were acquired using iVision software). Data are representative of >3 experiments from independent donors with similar results. (B) Cells were processed for TEM directly on slide surfaces. A23187-induced (2 μM, 60 minutes) EETosis eosinophils exhibit disrupted plasma membranes with clusters of released secretory granules (arrows) and DNA nets (Nets). Extracellular free granules (B, insert) show their typical morphologies with full crystalloid cores and matrices and intact granule-delimiting membranes. Some granules were entrapped in DNA nets. Scale bars represent 20 μm (A), 1 μm (B), and 700 nm (B, insert).
Intact granules and limited granule protein localization at extracellular DNA nets released through EETosis-mediated cell lysis. (A) Eosinophil EETosis was induced using 1 mg/mL immobilized IgG (120 minutes) or 2 μM A23187 (60 minutes) and fixed and permeabilized, then stained for MBP (green) and DNA (PI, red). Most released DNA nets did not contain free granule MBP protein and instead showed punctate or cell-associated staining. Images were obtained with a BX62 Olympus upright microscope, 20× UPlanApo objective with a numerical aperture of 0.70, coupled to a Hamamatsu Orca-AG fire-wire cooled digital camera (Hamamatsu Photonics, Hamamatsu, Japan; images were acquired using iVision software). Data are representative of >3 experiments from independent donors with similar results. (B) Cells were processed for TEM directly on slide surfaces. A23187-induced (2 μM, 60 minutes) EETosis eosinophils exhibit disrupted plasma membranes with clusters of released secretory granules (arrows) and DNA nets (Nets). Extracellular free granules (B, insert) show their typical morphologies with full crystalloid cores and matrices and intact granule-delimiting membranes. Some granules were entrapped in DNA nets. Scale bars represent 20 μm (A), 1 μm (B), and 700 nm (B, insert).
EETosis releases eosinophil granules both bound within plasma membranes and later as free granules following plasma membrane lysis
To study the EETosis processes that liberate intact eosinophil granules extracellularly, we monitored eosinophils with time-lapse, phase-contrast microscopy (Figure 5A, supplemental Video 1, and supplemental Video2). Eosinophils undergoing ETosis showed common morphological behaviors, starting with the arrest of their chemokinetic migration and with enhanced, rapid intracytoplasmic movements of their granules. As shown in supplemental Figure 4, the eosinophil’s bilobed nucleus progressively condensed into a single round shape. PM protrusions developed, and at times 1 to 3 granules were released together with enveloping PMs by budding off from the PM protrusions. In addition, larger collections of granules were released from eosinophils into PM-bound clusters. Thus, the budding release of small numbers of granules and the extrusion of larger clusters of granules resulted in the production of a variety of sizes of PM-bound, granule-packed structures. The release of granule structures typically precedes by two to five minutes nuclear membrane disintegration and nuclear chromatolysis. With the subsequent lysis of the eosinophils and the disruption of their PMs, the remaining intracellular granules were liberated.
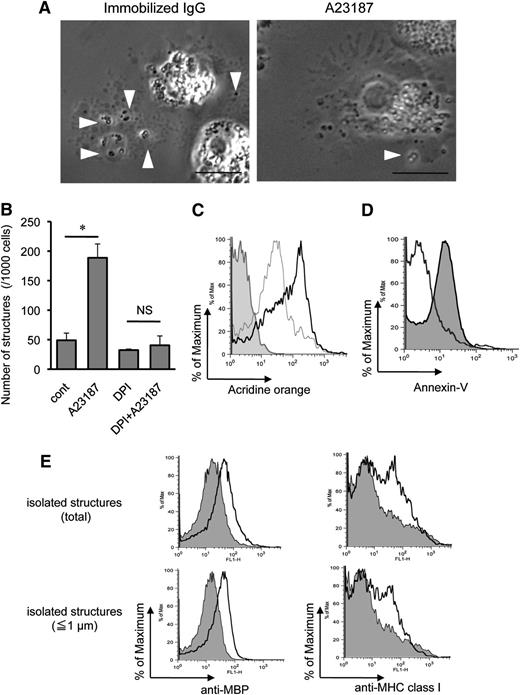
Characteristics of EETosis-induced intact eosinophil granule release. (A) Several cell-free granules were released by budding from PMs of eosinophils undergoing ETosis. Eosinophils were studied with time-lapse, phase-contrast imaging every 3 seconds following 1 mg/mL immobilized IgG or 2 μM A23187 stimulation. Images were captured from supplemental Video 1 and supplemental Video 2 (Nikon Eclipse TE300, 100× PlanApo objective with a numerical aperture of 1.40, equipped with a Qimaging Retiga EXi cooled digital camera in conjunction with iVision image analysis software). Eosinophils that initially showed a typical bilobed nucleus then demonstrated that peripheral small PM protrusions developed. Arrowheads show releasing/released granules. After nuclear membrane disintegration to release DNA nets within the cytoplasm, eosinophils gradually turned into a rounded shape, and granule movement slowed with deflected intracellular distribution of the granules. Several extracellular granules remained attached to the culture plate. Data representative of >3 experiments of independent donors with similar results are shown. (B) EETosis-elicited production of subcellular structures (⩽4 μm) was inhibited by DPI. Eosinophils were stimulated with A23187 (2 μM) in the presence of DPI in 0.1% BSA containing RPMI medium for 60 minutes. Subcellular structures in culture medium were counted as described in “Materials and methods” and supplemental Figure 2. Data (triplicate, mean ± SD) are representative of 3 experiments from independent donors with similar results. *P < .05. (C) Released cell-free structures retained the lysosomal dye AO. AO-loaded eosinophils were stimulated with A23187 for 1 hour. The structures (⩽4 μm) in culture medium from different preparations were subjected for flow cytometry. The graph shows the structures from AO-loaded cells (black line); fixed (2% PFO) and permeabilized (0.1% saponin) structures from AO-loaded cells (gray line); and structures from cells without AO staining (filled histogram). Data are representative of 3 experiments from independent donors with similar results. (D) A23187-induced subcellular structures were not apoptotic bodies. Subcellular structures (⩽4 μm) from A23187 and anti-Fas Ab–stimulated cells were stained with annexin V, followed by flow cytometric analysis. Fas-stimulated apoptotic cells produce annexin V–positive subcellular structures (ie, apoptotic bodies, filled histogram), but A23187-stimulated ETosis cells did not (open histogram). Apoptosis was induced, as described in supplemental Figure 4. Data are representative of 3 experiments from independent donors with similar results. (E) Isolated granule-rich subcellular structures retained the granule protein MBP; some were variably associated with PM-derived MHC class I proteins, regardless of size. Culture medium of A23187-stimulated cells was collected, and residual cells were removed by centrifugation (200 × g for 10 minutes). DNA was removed by DNase treatment, and buoyant vesicles and membranes were removed by further centrifugation (2500 × g for 10 minutes). Isolated granule-rich structures were stained for MBP and MHC class I (open histograms). Total structures (⩽4 μm) and small structures (⩽1 μm) were gated. Filled histograms show isotype-matched controls. Data are representative of 3 experiments from independent donors with similar results. Scale bars represent 5 μm (A). NS, not significant.
Characteristics of EETosis-induced intact eosinophil granule release. (A) Several cell-free granules were released by budding from PMs of eosinophils undergoing ETosis. Eosinophils were studied with time-lapse, phase-contrast imaging every 3 seconds following 1 mg/mL immobilized IgG or 2 μM A23187 stimulation. Images were captured from supplemental Video 1 and supplemental Video 2 (Nikon Eclipse TE300, 100× PlanApo objective with a numerical aperture of 1.40, equipped with a Qimaging Retiga EXi cooled digital camera in conjunction with iVision image analysis software). Eosinophils that initially showed a typical bilobed nucleus then demonstrated that peripheral small PM protrusions developed. Arrowheads show releasing/released granules. After nuclear membrane disintegration to release DNA nets within the cytoplasm, eosinophils gradually turned into a rounded shape, and granule movement slowed with deflected intracellular distribution of the granules. Several extracellular granules remained attached to the culture plate. Data representative of >3 experiments of independent donors with similar results are shown. (B) EETosis-elicited production of subcellular structures (⩽4 μm) was inhibited by DPI. Eosinophils were stimulated with A23187 (2 μM) in the presence of DPI in 0.1% BSA containing RPMI medium for 60 minutes. Subcellular structures in culture medium were counted as described in “Materials and methods” and supplemental Figure 2. Data (triplicate, mean ± SD) are representative of 3 experiments from independent donors with similar results. *P < .05. (C) Released cell-free structures retained the lysosomal dye AO. AO-loaded eosinophils were stimulated with A23187 for 1 hour. The structures (⩽4 μm) in culture medium from different preparations were subjected for flow cytometry. The graph shows the structures from AO-loaded cells (black line); fixed (2% PFO) and permeabilized (0.1% saponin) structures from AO-loaded cells (gray line); and structures from cells without AO staining (filled histogram). Data are representative of 3 experiments from independent donors with similar results. (D) A23187-induced subcellular structures were not apoptotic bodies. Subcellular structures (⩽4 μm) from A23187 and anti-Fas Ab–stimulated cells were stained with annexin V, followed by flow cytometric analysis. Fas-stimulated apoptotic cells produce annexin V–positive subcellular structures (ie, apoptotic bodies, filled histogram), but A23187-stimulated ETosis cells did not (open histogram). Apoptosis was induced, as described in supplemental Figure 4. Data are representative of 3 experiments from independent donors with similar results. (E) Isolated granule-rich subcellular structures retained the granule protein MBP; some were variably associated with PM-derived MHC class I proteins, regardless of size. Culture medium of A23187-stimulated cells was collected, and residual cells were removed by centrifugation (200 × g for 10 minutes). DNA was removed by DNase treatment, and buoyant vesicles and membranes were removed by further centrifugation (2500 × g for 10 minutes). Isolated granule-rich structures were stained for MBP and MHC class I (open histograms). Total structures (⩽4 μm) and small structures (⩽1 μm) were gated. Filled histograms show isotype-matched controls. Data are representative of 3 experiments from independent donors with similar results. Scale bars represent 5 μm (A). NS, not significant.
In other cells, activated cells can induce vesicle-shedding without affecting cell viability.34 To verify that the release of intact granules from eosinophils was associated with EETosis, we counted the absolute number of released subcellular structures (≤4 μm with high granularity) in the culture medium by using a flow cytometer (supplemental Figure 1). Notably, the release of extracellular free granule structures with A23187 was completely inhibited by DPI (Figure 5B). Enhanced release of granule structures was associated with increasing EETosis, as evidenced by concentration dependency (supplemental Figure 6A) and time course (supplemental Figure 6B). Moreover, extracellular structures were not liberated at 4°C (supplemental Figure 6C), indicating an active cellular process. To confirm the stability of released cell-free granules, we loaded eosinophil granules with AO, a lysosomal granule dye, followed by stimulation with A23187. After 1 hour, abundant cell-free, AO-retained granules were found in culture medium (supplemental Figure 7A). Consistent with Figure 5A, granules were found as single or grouped structures as well as PM-enveloped clusters containing different numbers of granules. Released subcellular structures were further subjected to flow cytometry; and in concert with fluorescent microscopy, most subcellular granule structures retained the granule dye (Figure 5C). Importantly, in contrast to apoptotic bodies derived from anti-Fas Ab–treated cells (supplemental Figure 3B), EETosis-induced subcellular structures lacked PS expression on their surfaces (Figure 5D).
To obtain granule-rich subcellular structures, any shed vesicles, membranes, and residual contaminating cells were removed by differential centrifugation. In concert with their identities as eosinophil granules, as shown in Figure 5E, isolated structures were positive for their contents of granule MBP. Moreover, there was bimodal staining for PM-derived major histocompatibility complex (MHC) class I protein, indicating that some, but not all, granule-containing structures lacked evidence of an associated PM. To target single granules (∼0.5 to1 μm in diameter), we also gated for small structures (≤1 μm), and similar results were obtained (Figure 5E, lower panels). Indeed, both PM-bound granules (supplemental Figure 7B,C) and PM-free granules (data not shown) were observed microscopically. To assess the intactness of PM-bound granules, live eosinophils were loaded with LysoTracker for labeling granules and CellTracker for cytoplasm and were stimulated with A23187. Intact granules were found both with and free of CellTracker-positive cytoplasm (supplemental Figure 7D). Similarly staining structures were observed following immobilized IgG- or PMA-induced EETosis (data not shown).
Collectively, we conclude that (1) EETosis releases eosinophil granules, (2) the extracellular granules retain specific granule proteins even after being released in culture medium, (3) some released granules may singularly or in clusters be surrounded by PS-nonexposed intact PM, and (4) some cytolytically released individual granules were devoid of PM.
EETosis-released granules free of plasma membranes are secretion competent and release ECP in response to CCL11
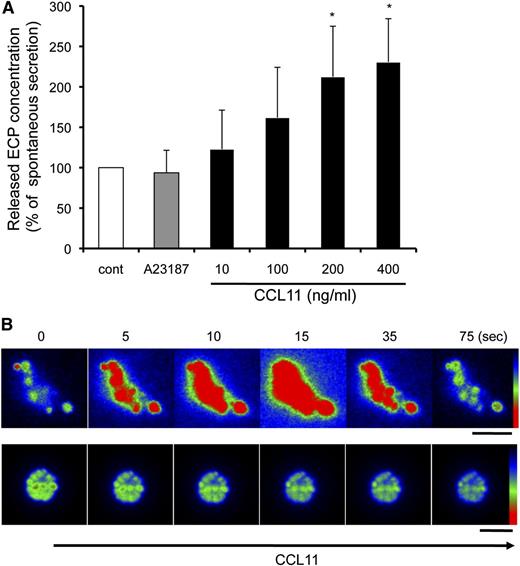
We previously demonstrated that human eosinophil granules, as isolated free of PMs principally by subcellular fractionation, express functional CCR3 chemokine receptors and secrete ECP in response to the ligand CCL11 by using their intragranular signaling and membranotubular network-based secretion systems.12,14 We evaluated whether EETosis-released granules would possess similar functional secretory properties. Following the stimulation of eosinophils with 2 μM A23187 for 90 minutes, ∼4% of the total ECP was recovered within the released granule structures. Isolated granules were stimulated with A23187 or CCL11 for 60 minutes, and secreted ECP was measured. Interestingly, the released granules did not respond to A23187. In contrast, the granules secreted granule-derived ECP in response to CCL11 in a concentration-dependent manner (Figure 6A).
Released granules were secretion competent in response to CCL11. (A) Culture medium of A23187-stimulated cells (2 μM, 90 minutes) was collected, and granule-rich subcellular structures were isolated. The cell-free structures were stimulated with 2 μM A23187 or the indicated concentrations of CCL11 for 60 minutes. Secreted ECP levels, assessed by enzyme-linked immunosorbent assay, were normalized with spontaneous ECP release (100%), and data are expressed as means ± SD, from 3 different donors. *P < .05 vs nonstimulated controls. The spontaneous secretion levels were 10.0 ± 4.4% of the total ECP in the structures. (B) AO-loaded eosinophils were stimulated with 2 μM A23187 to induce EETosis, followed by the isolation of subcellular granule structures. Among the 80 single granules or groups of granules, following CCL11 stimulation, significant responses with intense transient fluorescent flashes indicative of the release of monomeric AO were observed from 14 nonenveloped granules (17.5%). The secretory response was not observed by likely PM-bound clusters of granules (lower panels). Images were obtained with a Hamamatsu Orca-AG fire-wire cooled digital camera coupled to a BX62 Olympus microscope using a 60× PlanApo objective with a numerical aperture of 1.42. Fluorescence intensity was analyzed by iVision software and pseudocolored with red to represent the greatest intensity as indicated by the scale color. Experiments were repeated with eosinophils from 8 independent donors. The scale bar represents 3 μm (B).
Released granules were secretion competent in response to CCL11. (A) Culture medium of A23187-stimulated cells (2 μM, 90 minutes) was collected, and granule-rich subcellular structures were isolated. The cell-free structures were stimulated with 2 μM A23187 or the indicated concentrations of CCL11 for 60 minutes. Secreted ECP levels, assessed by enzyme-linked immunosorbent assay, were normalized with spontaneous ECP release (100%), and data are expressed as means ± SD, from 3 different donors. *P < .05 vs nonstimulated controls. The spontaneous secretion levels were 10.0 ± 4.4% of the total ECP in the structures. (B) AO-loaded eosinophils were stimulated with 2 μM A23187 to induce EETosis, followed by the isolation of subcellular granule structures. Among the 80 single granules or groups of granules, following CCL11 stimulation, significant responses with intense transient fluorescent flashes indicative of the release of monomeric AO were observed from 14 nonenveloped granules (17.5%). The secretory response was not observed by likely PM-bound clusters of granules (lower panels). Images were obtained with a Hamamatsu Orca-AG fire-wire cooled digital camera coupled to a BX62 Olympus microscope using a 60× PlanApo objective with a numerical aperture of 1.42. Fluorescence intensity was analyzed by iVision software and pseudocolored with red to represent the greatest intensity as indicated by the scale color. Experiments were repeated with eosinophils from 8 independent donors. The scale bar represents 3 μm (B).
The activation of AO-loaded eosinophil granules releases, into nonacidic media, monomeric AO that exhibits a spectral shift to green, yielding transient green flashes from secretion-responding granules.14 To obtain direct evidence showing that EETosis-released granules were responsive to CCL11, after AO-loaded cells were treated with 2 μM A23187, isolated granules were stimulated with CCL11. Some EETosis-released free granules exhibited rapid activation in response to CCL11, as evidenced by fluorescent flashes attributable to the release of AO (Figure 6B, upper panels). In contrast, some granules, including both free and those likely clustered within PMs, failed to exhibit secretory AO flashes in response to CCL11 (Figure 6B, lower panels). Stimulation of isolated granules with medium or A23187 did not induce AO secretion (data not shown). These results indicate that some granules extruded during A23187-mediated EETosis retained their capacity for CCL11-elicited secretory responses.
Discussion
Eosinophils as granulocytes contain distinct cytoplasmic granules, notable by their ultrastructurally unique crystalline cores and by their content of proteins, including specific cationic proteins as well as preformed cytokines. In diverse human diseases in which eosinophils may participate, there have been recognitions over many years that cell-free eosinophil granules, released from eosinophils, are found in tissues or secretions.4 These findings include ultrastructural studies recognizing intact free eosinophil granules;8,10 immunofluorescent demonstrations of eosinophil cationic proteins in extracellular granule foci; and, especially in asthma, clusters of extracellular eosinophil granules.7 Disruptions of PMs and nuclear membranes along with chromatolysis have been observed in eosinophils associated with allergic and anthelmintic responses.28,29 Our studies aimed to examine the mechanisms and consequences by which human eosinophils could cytolytically and extracellularly release their intact granules.
Ultrastructural examination of eosinophils in the sinus tissue of a patient with allergic sinusitis and the skin of a patient with a hypereosinophilic syndrome, respectively, (Figure 1) revealed the liberation of free extracellular granules as well as nuclear changes (disruption of nuclear membranes and decondensation of nuclear chromatin) that were distinct from apoptosis or necrosis. Apoptotic eosinophils exhibit condensed compaction of chromatin against the nuclear envelope and shrinkage and budding of the whole cell to form membrane-bound, PS-exposed apoptotic bodies. Neither anti-Fas–elicited eosinophil apoptosis nor heat-elicited eosinophil necrosis yielded extracellular release of nuclear DNA or cell-free eosinophil granules (supplemental Figure 3). To investigate mechanisms of eosinophil cytolysis, we used immobilized immunoglobulins (IgG, IgA), PAF with either IL-5 or GM-CSF, and nonphysiologic stimuli, calcium ionophore A23187 and PMA. Each of these stimuli elicited eosinophil lytic death (EETosis) that was distinct from apoptosis or necrosis. EETosis occurred progressively over 30 to 120 minutes and was dependent on the generation of ROS, being inhibitable by DPI. These stimuli are known to induce/prime ROS production by eosinophils.35-37
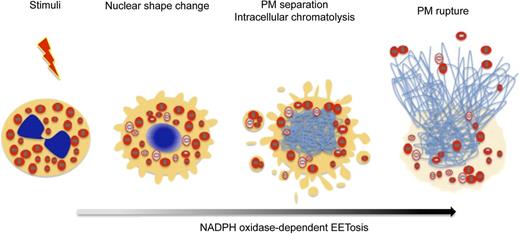
Prior studies demonstrated that cytokine-primed human eosinophils, in response to certain agonists such as lipopolysaccharide, C5a, and CCL11, can rapidly (within seconds) and noncytolytically release not nuclear, but mitochondrial DNA to form extracellular DNA nets.21 Our findings identify another means by which eosinophils can form DNA nets, more akin temporally and mechanistically to ETosis in neutrophils16 and mast cells.17 As in neutrophils,18 eosinophil ETosis involves a more delayed, cytolytic release of nuclear DNA to form extracellular DNA nets. The temporal sequence of changes within eosinophils undergoing cytolysis is presented in Figure 7. Following stimuli-elicited initiation of eosinophil cytolysis, the typically bilobed nuclei of eosinophils lost their lobulation to coalesce into single nuclear structures. During this time, some eosinophil granules were released extracellularly as PM-enveloped structures. Thereafter, nuclei disintegrated intracellularly to form intracellular DNA nets. Subsequently, the eosinophils’ PMs rupture, releasing both nuclear DNA-derived (histone-positive) nets and free eosinophil granules. Eosinophil ETosis-mediated cytolysis leads to the extracellular liberation of intact eosinophil granules.
EETosis induces the release of intact cell-free granules through budding and cell lysis. The graphic shows the temporal course of the morphologic changes exhibited by eosinophils undergoing EETosis. Stimuli-elicited NADPH oxidase activation leads to the common process of eosinophil cytolysis, followed by the loss of the typically bilobed nuclei into a single round nuclei. During this time, some eosinophil granules were released extracellularly as PM-enveloped structures. Thereafter, nuclei disintegrated to form intracellular DNA nets. Subsequently, the eosinophils’ PMs rupture, releasing both a weblike chromatin structure and free eosinophil granules. EETosis-mediated cytolysis leads to the extracellular liberation of intact granules, some of which retained their capacity for secretory responses.
EETosis induces the release of intact cell-free granules through budding and cell lysis. The graphic shows the temporal course of the morphologic changes exhibited by eosinophils undergoing EETosis. Stimuli-elicited NADPH oxidase activation leads to the common process of eosinophil cytolysis, followed by the loss of the typically bilobed nuclei into a single round nuclei. During this time, some eosinophil granules were released extracellularly as PM-enveloped structures. Thereafter, nuclei disintegrated to form intracellular DNA nets. Subsequently, the eosinophils’ PMs rupture, releasing both a weblike chromatin structure and free eosinophil granules. EETosis-mediated cytolysis leads to the extracellular liberation of intact granules, some of which retained their capacity for secretory responses.
In neutrophils undergoing ETosis, granule-contained proteins can associate intracellularly with nuclear DNA even before PMs rupture, and granule proteins are bound to extracellular DNA nets.19,38 Intact neutrophil granules are not recognized as bound to extracellular DNA nets. In contrast, in eosinophils undergoing ETosis, granules are released as intact structures. Our findings indicated that eosinophil granule proteins were not predominantly free and bound to DNA nets, but rather they remained principally within granules that themselves could bind to DNA nets. Moreover, we showed that some released cell-free eosinophil granules exhibited secretion competency in response the eosinophil-active chemokine CCL11 and secreted ECP and granule AO (Figure 6). Other granules, including clusters of likely PM-enveloped granules, did not exhibit individual CCL11-elicited granule secretory responses, at least in short-term in vitro assays. Whether the clusters of PM-enveloped granules might exhibit secretory competence in vivo requires further study.
In conclusion, our results reveal a process by which eosinophils undergo cytolysis to liberate intact cell-free granules, as is well recognized in tissues and secretions in association with diverse eosinophil-associated diseases. The process of ETosis in eosinophils is akin to that in neutrophils18 but notably differs in that intact eosinophil granules are released extracellularly and still contain the bulk of their protein contents. At least some EETosis-released granules remain secretion competent. Thus, EETosis results in the generation of histone-bearing nuclear DNA extracellular nets and cell-free granules, both of which may exert biological activities for eosinophils postmortem.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful to Kristen Young and Jason Xenakis for their outstanding technical assistance and Dr. Praveen Akuthota and Professor Hélio Chiarini-Garcia for helpful discussions.
This work was supported in part by Uehara Memorial Foundation (S.U.); Conselho Nacional de Desenvolvimento Científico e Tecnológicoand Fundação de Amparo à Pesquisa do Estado de Minas Gerais (R.C.N.M.); National institute of Health: National Heart, Lung and Blood Institute R01-HL096795 (I.G.); R01-HL095699 (L.A.S.); and grants from the National Institutes of Health: National Institute of Allergy and Infectious Diseases (R37-AI020241 and R01-AI051645, P.F.W.).
National Institutes of Health
Authorship
Contribution: S.U. designed and performed experiments and wrote the manuscript. R.C.N.M. and A.M.D. performed electron microscopic experiments and wrote the manuscript. I.G. provided technical assistance and edited the manuscript. L.A.S. contributed scientific advice and discussions. P.F.W. supervised the research and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Peter F. Weller, MD, 330 Brookline Ave, CLS-943, Boston, MA 02215; e-mail: pweller@bidmc.harvard.edu.