Abstract
Anemia linked to a relative deficiency of renal erythropoietin production is a significant cause of morbidity and medical expenditures in the developed world. Recombinant erythropoietin is expensive and has been linked to excess cardiovascular events. Moreover, some patients become refractory to erythropoietin because of increased production of factors such as hepcidin. During fetal life, the liver, rather than the kidney, is the major source of erythropoietin. In the present study, we show that it is feasible to reactivate hepatic erythropoietin production and suppress hepcidin levels using systemically delivered siRNAs targeting the EglN prolyl hydroxylases specifically in the liver, leading to improved RBC production in models of anemia caused by either renal insufficiency or chronic inflammation with enhanced hepcidin production.
Introduction
In adult mammals, the kidney is the major source of the hormone erythropoietin, which stimulates red blood cell (RBC) production. As a result, patients with chronic renal failure are often anemic due at least partly to inadequate erythropoietin (EPO) production. Treatment with pharmacologic doses of recombinant EPO is expensive and has recently been linked to increased morbidity and mortality due to cardiovascular events.1,2 Some patients also become refractory to recombinant erythropoietin over time because of a variety of host factors, including Abs against the recombinant protein itself or altered iron homeostasis secondary to humoral factors such as hepcidin.3,4
Transcription of the erythropoietin gene (EPO) is controlled by the heterodimeric transcription factor hypoxia-inducible factor (HIF) and is therefore intimately linked to oxygen delivery to the kidneys, which are normally borderline hypoxic at rest and poised to respond to further decrements in oxygen delivery.5 HIF consists of an unstable α-subunit (eg, HIF1α or HIF2α) and a stable β-subunit (eg, HIF1β, which is also called ARNT1).6 In the presence of oxygen, HIFα becomes prolyl hydroxylated by members of the EglN (also called PHD) family of 2-oxoglutarate–dependent dioxygenases, leading to its polyubiquitination and proteasomal degradation.6 As oxygen levels decrease, EglN activity is diminished, leading to HIF stabilization and activation. There are 3 EglN family members: EglN1 (also called PHD2) is the primary regulator of HIF and EglN2 (PHD1) and EglN3 (PHD3) play compensatory roles under certain circumstances.6 The analysis of families, as well as of genetically engineered mice, with hereditary polycythemia (excess RBCs) implicate EglN1 and HIF2α as the critical regulators of erythropoietin, and thus RBC production, in adults.7,8
During fetal life, the liver, rather than the kidney, is the major source of erythropoietin. Moreover, nonrenal tissues, most likely including the liver, can contribute to erythropoietin production in anephric, hypoxemic adults.9-11 We showed recently that erythropoietin production was markedly induced in adult hepatocytes lacking all 3 EglN family members, supporting the idea that the liver can contribute to circulating erythropoietin levels in the setting of chronic kidney disease.12
Orally available EglN inhibitors have been shown to stimulate RBC production in preclinical models and are currently being tested in the clinical setting as a potential treatment for anemia, including anemia linked to chronic kidney disease.11,13,14 However, it is not yet known whether chronic, systemic HIF activation will lead to untoward side effects in humans. Moreover, it is possible that some EglN inhibitors will have off-target effects such as inhibiting other enzymes that are structurally related to the EglN family members. In an effort to achieve a more targeted approach, in the present study, we exploited the ability of lipid nanoparticles (LNPs) to deliver siRNAs specifically to the liver as a means of inactivating hepatic EglN activity.
Methods
Cre-Lox experiments
EglN1 flox/flox (EglN1f/f) and HIF1α-Luc reporter mice were described previously.13,15 The EglN2−/− and EglN3−/− mice were a gift of Regeneron Pharmaceuticals. All of the EglN strains were backcrossed to C57BL/6 mice at least 6 times. Four- to 8-week-old EglN1f/fEglN2−/−EglN3−/− and EglN1f/fEglN2+/+EglN3+/+ mice were used. Adenoviruses carrying GFP (Ad5CMVeGFP) or GFP-Cre (Ad5CMVCre-eGFP; 2 × 109 PFU/200 μL) were obtained from Gene Transfer Vector Core, University of Iowa and were administered to mice by tail vein injection.
siRNA design, synthesis, and screening
Mouse reference sequences for each EglN gene, EglN1-NM_022051.2, EglN2-NM_053046.2, and EglN3-NM_022073.3, were used for siRNA selection. To identify corresponding siRNAs with low potential for cross-reactivity with irrelevant targets (off-targets) 19mer candidate sequences were subjected to a homology search against the RefSeq mRNA database. A set of off-target properties, number of mismatches in nonseed region, number of mismatches in seed region, and number of mismatches in cleavage site region were extracted for each 19mer to calculate an off-target score to rank the most specific siRNAs against each EglN target for synthesis. Single-stranded RNAs were synthesized at Alnylam Pharmaceuticals on controlled-pore glass using standard phosphoramidite chemistry. The phosphoramidites were obtained from Proligo Biochemie. Deprotection and purification of the crude oligoribonucleotides was done by ion exchange HPLC according to established procedures.16 Yield and concentration were determined by UV absorption of the respective RNA at 260 nm using a spectral photometer (Beckman-Coulter). siRNA was generated by mixing equimolar amounts of complementary strands in annealing buffer (50nM sodium acetate, pH 5.5), heating in a water bath at 90°C for 3 minutes, and cooling to room temperature over several hours.
Mouse hepatocyte BNL-C12 cells cultured in MEM (Invitrogen) supplemented with 10% FBS, penicillin (100 units/mL), and streptomycin (100 μg/mL) were transfected with siRNA targeting EglN1, EglN2, or EglN3 using RNAiMAX (Invitrogen). Twenty-four hours after transfection, cells were lysed and the respective EglN target mRNA levels compared with a housekeeping control were measured with a branched-DNA assay according to the manufacturer's instructions (Panomics). Normalized ratios of EglN mRNA to housekeeping controls were compared with control siRNA-transfected cells to identify potent siRNAs against each target for in vivo studies. (For sequences, please see supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article.)
siRNA formulation in LNPs
The LNPs were prepared with an ionizable lipid, disteroylphosphatidyl choline, cholesterol, and PEG-DMG using a spontaneous vesicle formation procedure as previously described at a component molar ratio of ∼ 50/10/38.5/1.5.17,18 The final lipid: siRNA ratio was ∼ 12:1. The particle size of LNPs was determined by dynamic light scattering (Zetasizer Nana ZS; Malvern) and the mean diameter was in the range of 40-70 nm for all LNPs used in these studies. The siRNA entrapment efficiency was determined by Ribogreen assay to be > 95%.
LNP dosing and anemia models
All procedures used in animal studies were approved by the Alnylam Pharmaceuticals institutional animal care and use committee and were consistent with local, state, and federal regulations. Wild-type C57BL/6 or 5/6th nephrectomized FVB/N mice or Sprague Dawley rats mice received either PBS or LNP formulations via tail vein injection at a volume of 0.01 mL/g. For blood draws animals were anesthetized by isoflurane inhalation and blood was collected into serum separator tubes or EDTA-containing tubes by retroorbital bleeds. In some cases tail pricks were used for small amounts of blood to monitor hematocrit levels using capillary tubes. For 5/6 nephrectomy experiments animals underwent the procedure at Charles River Laboratories. One kidney was removed and the animal was allowed to recover 1 week before 2/3 of the second kidney was removed. Over 2-3 weeks the hematocrit levels were monitored every 5 days to determine anemia progression. Treatment began when hematocrit levels reached approximately 30%. Animals were dosed every 4 days for a total of 3 doses and killed 4 days after the final dose for hematology and tissue mRNA analysis. For rat anemia experiments, a single IP injection of PG-APS (a polymer derived from group A streptococci) was used to induce arthritis.19-21 After an acute phase of inflammation the rats were bled weekly for CBC analysis, with chronic anemia (hematocrit < 35%) developing by day 21. Beginning at day 21 animals were dosed with siRNA weekly for 3 total doses with serum and hematology parameters measured bi-weekly throughout the treatment period. Animals were killed at day 37 and tissues harvested to measure target mRNA levels.
Serum Epo ELISA and hematology
At various time points or immediately before animal sacrifice blood was collected and processed to serum (Microcontainer serum separator tubes; Becton Dickinson). Serum Epo was measured using a mouse/rat Erythropoietin ELISA kit (R&D Systems) according to the manufacturer's instructions. Standard complete blood counts (CBC) were monitored using an Advia120 system (Bayer).
Tissue mRNA quantification
After animals were killed tissues were snap frozen in liquid nitrogen and ground into powders. Tissue lysates were prepared in lysis buffer containing Proteinase K (Epicenter) and levels of siRNA target mRNA determined relative to GAPDH or Beta-Actin housekeeping genes using a branched DNA assay according to the manufacturer's instructions (Quantigene reagent system; Panomics).
5′RACE assay
Total RNA was isolated from sorted cells using Trizol (Invitrogen) and RNAeasy mini columns (QIAGEN). Roughly 2μg of RNA was used for 5′RACE similar to the Generacer kit (Invitrogen) protocol. The oligonucleotide adaptor was ligated directly to total RNA using T4 RNA ligase (2U) for 1 hour at 37°C. The ligation mixture was reverse transcribed using the EglN1, EglN2, and EglN3 gene-specific oligos (supplemental Table 2) using Superscript III (Invitrogen) for 1 hour at 50°C, then 15 minutes at 70°C. cDNA was amplified using 2 μL of cDNA amplified with Platinum Taq (Invitrogen) for 35 cycles where Tm = 50°C with first round PCR EglN oligos paired with the adaptor oligo in individual reactions in a 50 μL total reaction volume. A second round of nested PCR was performed with 3 μL of the first reaction for 25 cycles where Tm = 55°C using the respective nested oligos and the nested adaptor oligo. PCR products were examined by gel electrophoresis, and the total PCR reaction was cloned into pCR4-TOPO vector for sequencing (Invitrogen).
Statistical analysis
Data shown are mean ± SD. Results were analyzed for statistical significance using 1- or 2-way ANOVAs where appropriate followed by Bonferroni multiple comparison posttests. Significance values shown (P < .05, .01, or .001) compare the treatment group to the corresponding Luciferase control.
Results
Acute EglN inactivation promotes hepatic Epo production
We recently showed that hepatic Epo production was preserved in adult mice in which all 3 EglN family members were genetically disrupted, either in the germ line or during late embryogenesis, but not in mice that retained at least one of the 3 family members.12 Moreover, we showed that acute inactivation of EglN1 in adult mice lacking EglN2 and EglN3 reactivated hepatic Epo production.12 These results left open the possibility that acute inactivation of EglN1 in mice that retained EglN2 and EglN3 would also promote hepatic Epo production in adult mice. To test this we administered an adenovirus encoding Cre recombinase (or control virus) by tail vein injection, which results in viral infection primarily in the liver,22 to EglN1f/fEglN2+/+EglN3+/+ mice and to EglN1f/fEglN2−/−EglN3−/− mice. Acute inactivation of EglN1 in the former increased circulating Epo and hematocrit levels although not to the degree achieved in mice that also lacked EglN2 and EglN3 (supplemental Figure 1). Moreover, hepatic Epo mRNA levels were persistently elevated in livers lacking all 3 EgN paralogs whereas they reverted toward normal in mice that retained EglN2 and EglN3 (supplemental Figure 1). Therefore acute inactivation of EglN1 stimulates hepatic Epo production but this effect is transient in nature because of compensation by EglN2 and/or EglN3. Simultaneous inactivation of all 3 EglN family members appears to be necessary and sufficient for sustained, high level, hepatic Epo production in genetically engineered models of EglN inactivation.
Consistent with this, pan-EglN small molecule inhibitors induce Epo/EPO production in anephric mice and in patients with chronic renal failure.11,13,14 These agents, however, might theoretically affect other 2-oxoglutarate–dependent dioxygenases and affect tissues other than the liver. In an effort to achieve a more targeted approach, we exploited the fact that many currently available siRNA delivery methods preferentially target the liver.17,23,24 Significant improvements in potency with the newest generation of LNPs have enabled robust knockdown of multiple genes simultaneously.18,25
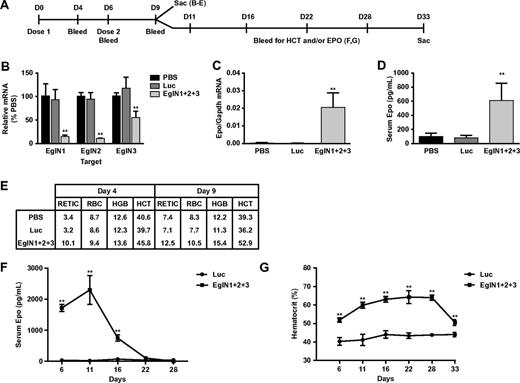
We first designed and validated siRNAs that are specific for mouse EglN1, EglN2, or EglN3, which were then administered by tail vein injection, either alone or in combinations using LNPs (Figure 1A). These siRNAs achieved dose dependent knockdown of their intended targets in vivo (Figure 1B and supplemental Figures 2-3). Knockdown of EglN3 mRNA in the setting of combined EglN inactivation was modest compared with treatment with EglN3 siRNA alone (Figure 1B and supplemental Figures 2-3), presumably because EglN3 mRNA is robustly induced by EglN1 loss.26 In keeping with our earlier results using Cre-Lox technology, inactivation of EglN1 alone led to a modest induction of circulating Epo (supplemental Figures 3-4), without a sustained increase in hepatic Epo mRNA production (supplemental Figures 3-4), whereas hepatic Epo mRNA (Figure 1C) and circulating Epo (Figure 1D) were dramatically increased in mice that received LNPs containing siRNAs against all 3 EglN family members. The induction of circulating Epo in this setting was detectable out to 2 weeks after siRNA administration (Figure 1F) and was associated with a sustained increase in RBC production (Figure 1E,G). Similar results were obtained with mixtures of 3 LNPs that contained single siRNAs against either EglN1, EglN2, or EglN3 (data not shown). Hematocrit values began to fall approximately 3 weeks after the last dose of EglN siRNA (approximately 2 weeks after the peak in serum Epo) with kinetics compatible with a normal murine RBC half-life of approximately 10 days27,28 (Figure 1F-G). Liver histology and serum transaminase and bilirubin levels remained normal in mice that were treated weekly with the EglN siRNA nanoparticles for 1 month before necropsy, despite robust induction of HIF1α and HIF2α (supplemental Figure 5 and data not shown).
EglN siRNA activates hepatic Epo production and stimulates erythropoiesis. (A) Overview of dosing schedule and bleeds. Mice were dosed intravenously with LNPs containing an equal mixture of 3 siRNAs targeting EglN1, EglN2, and EglN3, respectively (total dose, 1 mg/kg). Mice treated with PBS or a similarly prepared LNP containing a single siRNA against firefly luciferase served as controls. (B-D) Hepatic EglN mRNA (B), hepatic Epo mRNA (C) at day 9, and serum Epo (D) levels at day 4. mRNA levels were normalized to GAPDH mRNA levels and then to the corresponding values in PBS-treated mice. (E) Hematology measurements at day 4 after first dose or day 9 after 2 doses. (F-G) Serum Epo (F) and hematocrit (G) levels in mice treated as in panel A with 2 doses of siRNA against the EglN family members or against firefly luciferase (LUC). For panels A through E, n = 5; for panels F and G, n = 3. Error bars represent 1 SD. **P < .01.
EglN siRNA activates hepatic Epo production and stimulates erythropoiesis. (A) Overview of dosing schedule and bleeds. Mice were dosed intravenously with LNPs containing an equal mixture of 3 siRNAs targeting EglN1, EglN2, and EglN3, respectively (total dose, 1 mg/kg). Mice treated with PBS or a similarly prepared LNP containing a single siRNA against firefly luciferase served as controls. (B-D) Hepatic EglN mRNA (B), hepatic Epo mRNA (C) at day 9, and serum Epo (D) levels at day 4. mRNA levels were normalized to GAPDH mRNA levels and then to the corresponding values in PBS-treated mice. (E) Hematology measurements at day 4 after first dose or day 9 after 2 doses. (F-G) Serum Epo (F) and hematocrit (G) levels in mice treated as in panel A with 2 doses of siRNA against the EglN family members or against firefly luciferase (LUC). For panels A through E, n = 5; for panels F and G, n = 3. Error bars represent 1 SD. **P < .01.
Noninvasive pharmacodynamic imaging of EglN siRNA
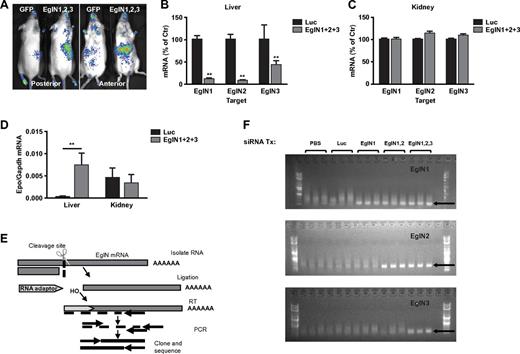
We previously showed that EglN activity can be monitored noninvasively in mice that ubiquitously express a HIF1α-luciferase fusion protein that contains a region of HIF1α that is sufficient to be hydroxylated by EglN and subsequently ubiquitinated by the pVHL ubiquitin ligase complex.13 As expected, administration of the EglN siRNA mix to these mice decreased hepatic, but not renal, EglN activity as determined by increased photon emission in the region of the liver, but not kidneys, after luciferin administration (Figure 2A). Branched DNA analysis confirmed that EglN1, EglN2, and EglN3 mRNAs were decreased in the liver (Figure 2B), but not in the other organs examined, such as the kidney (Figure 2C, supplemental Figure 6, and data not shown), and was associated with increased hepatic, but not renal, Epo mRNA production (Figure 2D). Epo mRNA induction was undetectable in spleen or any other tissue examined further supporting the hepatic origin of the serum Epo rise (data not shown). The EglN mRNA silencing primarily in liver is consistent with the known biodistribution of this class of LNPs.17,18 EglN knockdown did not increase HIFα mRNA levels, which is consistent with the finding that regulation of HIF by EglN is largely posttranscriptional (supplemental Figure 7).6
LNP-mediated EglN siRNA delivery is liver specific and down-regulates EglN via an RNAi mechanism. (A) Bioluminescent images of HIF1α-Luc mice 72 hours after a single intravenous dose of LNPs targeting all 3 EglN family members or, as a negative control, green fluorescent protein (GFP). Total dose was 1 mg/kg (0.33 mg/kg per family member). (B-C) Quantification of EglN mRNA levels in livers (B) and kidneys (C) of mice 1 week after treatment with LNPs targeting all 3 EglN family members or firefly luciferase. mRNA levels were normalized to GAPDH mRNA and then to corresponding values in mice treated with luciferase siRNA. (D) Quantification of Epo mRNA levels in liver and kidney in mice treated as in panels B and C. (E) Overview of 5′RACE assay to monitor the cleavage site of target mRNA. (F) 5′RACE of mouse liver total RNA isolates from mice treated with the indicated siRNAs. Specific cleavage sites were confirmed by sequencing of excised bands from gel. For panels B through D, n = 5; for panel F, n = 3. Error bars represent 1 SD. **P < .01.
LNP-mediated EglN siRNA delivery is liver specific and down-regulates EglN via an RNAi mechanism. (A) Bioluminescent images of HIF1α-Luc mice 72 hours after a single intravenous dose of LNPs targeting all 3 EglN family members or, as a negative control, green fluorescent protein (GFP). Total dose was 1 mg/kg (0.33 mg/kg per family member). (B-C) Quantification of EglN mRNA levels in livers (B) and kidneys (C) of mice 1 week after treatment with LNPs targeting all 3 EglN family members or firefly luciferase. mRNA levels were normalized to GAPDH mRNA and then to corresponding values in mice treated with luciferase siRNA. (D) Quantification of Epo mRNA levels in liver and kidney in mice treated as in panels B and C. (E) Overview of 5′RACE assay to monitor the cleavage site of target mRNA. (F) 5′RACE of mouse liver total RNA isolates from mice treated with the indicated siRNAs. Specific cleavage sites were confirmed by sequencing of excised bands from gel. For panels B through D, n = 5; for panel F, n = 3. Error bars represent 1 SD. **P < .01.
5′-RACE analysis using liver samples from mice treated with EglN siRNA in LNPs was done to confirm that the decrease in EglN mRNA levels was occurring by an RNAi mechanism and not because of off-target effects. The EglN transcripts 1, 2, and 3 were cleaved at the predicted site recognized by the corresponding siRNA sequence. After PCR amplification, the products run on a gel were of the expected size for each EglN gene and sequencing confirmed the predicted siRNA recognition sites (Figure 2E-F). We also observed an increase in hepatic photon emission in HIF1α-luciferase reporter mice treated with EglN1 siRNA alone, but not with EglN2 or EglN3 siRNA alone, consistent with EglN1 being the primary regulator of HIF1α under normal conditions (supplemental Figure 8).
EglN siRNA ameliorates anemia in preclinical models
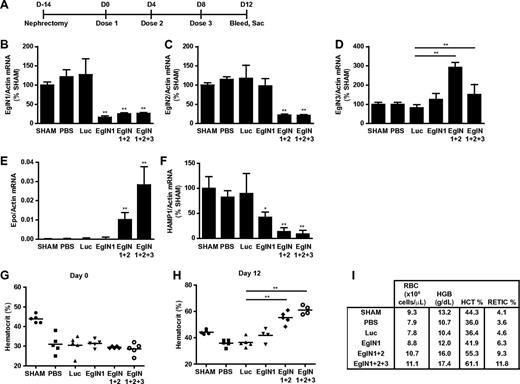
We also investigated whether EglN siRNA could be used to treat anemia in the setting of chronic renal failure. Mice were subjected to 5/6 nephrectomy, which is a widely used model for anemia linked to renal failure, or sham operations (Figure 3A,G). After developing anemia, the mice that had undergone nephrectomy were randomized to receive PBS, control siRNA (luciferase siRNA), siRNAs (singly or in combination) targeting EglN1, both EglN1 and EglN2, or all 3 EglN family members. Knockdown of the intended EglN mRNAs was again documented by branched-chain DNA analysis (Figure 3B-D). Consistent with the data in supplemental Figures 1, 3, and 4 described above, inactivation of EglN1 led to a modest increase in RBC production, which was markedly accentuated by coinactivation of EglN2 (Figure 3H-I). The maximal erythropoietic response, however, was observed after treatment with siRNA targeting all 3 EglN paralogs (Figure 3E,H-I). EglN inactivation in this model also decreased hepcidin mRNA levels, which is consistent with earlier studies using chemical hydroxylase inhibitors (Figure 3F).29,30 Control of hepcidin by HIF appears to be multifactorial and complex, potentially involving cell-autonomous effects of hepatic HIF on hepcidin transcription and indirect effects stemming from enhanced erythropoiesis.29,31-40
Targeting of EglN genes rescues anemia caused by renal failure. (A) Overview of 5/6 nephrectomy procedure and dosing schedule. (B-F) mRNA values at day 12 in mice treated with the indicated siRNAs as depicted in panel A. HAMP1 indicates hepcidin antimicrobial peptide 1. mRNA levels were normalized to actin mRNA and then to the corresponding sham mRNA level. Sham mice underwent sham surgery rather than 5/6 nephrectomy. (G-I) Baseline hematocrit (day 0; G), day 12 hematocrit (H), and day 12 hematology parameters (I) in mice treated with the indicated siRNAs as depicted in panel A (n = 5). Error bars represent 1 SD. *P < .05; **P < .01.
Targeting of EglN genes rescues anemia caused by renal failure. (A) Overview of 5/6 nephrectomy procedure and dosing schedule. (B-F) mRNA values at day 12 in mice treated with the indicated siRNAs as depicted in panel A. HAMP1 indicates hepcidin antimicrobial peptide 1. mRNA levels were normalized to actin mRNA and then to the corresponding sham mRNA level. Sham mice underwent sham surgery rather than 5/6 nephrectomy. (G-I) Baseline hematocrit (day 0; G), day 12 hematocrit (H), and day 12 hematology parameters (I) in mice treated with the indicated siRNAs as depicted in panel A (n = 5). Error bars represent 1 SD. *P < .05; **P < .01.
Chronic inflammation can lead to anemia due, at least in part, to increased levels of hepcidin and altered iron trafficking (anemia of chronic disease).41,42 Rats with experimental arthritis induced by PG-APS have been used as a model for the anemia linked to inflammation.19-21 In the 5/6 nephrectomy model, combined inactivation of EglN1 and EglN2 was sufficient to induce a brisk erythropoietic response (Figure 3E,H-I) and we were able to identify siRNAs that can effectively target rat EglN1 and EglN2 (Figure 4A-C). Treatment of anemic PG-APS rats with mixtures of siRNAs targeting both EglN1 and EglN2 decreased hepcidin levels (Figure 4D), increased hepatic Epo mRNA levels (Figure 4E), and corrected anemia (Figure 4F-G). Because treatment with EglN siRNA down-regulated hepcidin, a master regulator of iron homeostasis, we also examined several serum iron parameters. We saw trends of improvement in serum iron, ferritin, and transferrin saturation without significant effects on total iron-binding capacity (TIBC) or unsaturated iron-binding capacity (UIBC; supplemental Figure 9).
Targeting of EglN genes corrects anemia related to inflammation. (A) Overview of rat anemia of inflammation model and dosing schedule. Rats were dosed with PG-APS polymer or PBS on day 0. Rats treated with PG-APS developed anemia by day 21 and were then randomized to receive LNPs targeting both EglN1 and EglN2, LNPs targeting firefly luciferase (LUC), or not treated (PG-APS only). (B-E) Hepatic mRNAs levels at termination of study on day 37. mRNA levels were normalized to GAPDH levels and then to the corresponding value for mice that received PBS instead of PG-APS. (F-G) Hemoglobin (F) and hematocrit (G) values for rats treated with the indicated siRNAs as in panel A. n = 4 except for PBS (n = 3). Error bars represent 1 SD. *P < .05; **P < .01.
Targeting of EglN genes corrects anemia related to inflammation. (A) Overview of rat anemia of inflammation model and dosing schedule. Rats were dosed with PG-APS polymer or PBS on day 0. Rats treated with PG-APS developed anemia by day 21 and were then randomized to receive LNPs targeting both EglN1 and EglN2, LNPs targeting firefly luciferase (LUC), or not treated (PG-APS only). (B-E) Hepatic mRNAs levels at termination of study on day 37. mRNA levels were normalized to GAPDH levels and then to the corresponding value for mice that received PBS instead of PG-APS. (F-G) Hemoglobin (F) and hematocrit (G) values for rats treated with the indicated siRNAs as in panel A. n = 4 except for PBS (n = 3). Error bars represent 1 SD. *P < .05; **P < .01.
Discussion
The results of the present study suggest that systemically administered siRNAs targeting the EglN family would ameliorate anemias characterized by an absolute or relative deficiency of erythropoietin, such as anemias linked to chronic kidney disease or inflammation in humans. This approach would allow the body to produce native erythropoietin, thereby obviating the need for recombinant versions of this hormone. Moreover, other hepatic changes induced by EglN inhibition, such as decreased production of hepcidin, might enhance the effectiveness of endogenous erythropoietin and thereby lower the circulating erythropoietin levels needed to promote RBC production. This might be desirable if some of the cardiovascular complications of chronic erythropoietin production are more tightly linked to circulating erythropoietin levels, especially when supraphysiological, than to RBC mass per se.43
Small-molecule EglN inhibitors that interfere with the ability of these enzymes to use 2-oxoglutarate, iron, or both are currently in clinical development and are capable of stimulating erythropoietin production in the setting of chronic renal failure.11,13,14 In addition to those outlined in the previous paragraph for EglN siRNA, these agents have several theoretical advantages over recombinant erythropoietin, including an ability to activate many HIF-responsive genes that play roles in iron metabolism and erythropoiesis in tissues such as the liver, intestine, and BM. Therefore, EglN inhibitors may mimic the normal adaptation to hypoxemia more faithfully than do pharmacologic doses of erythropoietin. However, it is possible that some small-molecule EglN inhibitors will have off-target toxicities related to inhibition of other 2-oxoglutarate–dependent dioxygenases. Moreover, hypothetical concerns have been raised regarding the potential risks of chronic, systemic HIF activation.44,45 The theoretical targeting precision of siRNA, coupled with the ability to restrict delivery to the organ of interest (liver), might therefore translate into an advantage for EglN siRNA compared with small-molecule EglN inhibitors in the clinical setting.
Null alleles for targets of interest are often used to predict the consequences of inhibiting those targets in patients with drug-like small molecules. However, germline-null alleles can be impractical or misleading if their protein products also play roles during embryological development. This problem can be circumvented by the use of conditional alleles that can be inactivated using, for example, Cre-Lox technology. The loss of alleles produced by such recombination events, however, typically mimic complete and irreversible target loss, whereas most successful drugs are useful because they induce partial, reversible target inhibition that can be titrated to achieve their desired effects. Based on these considerations, one might predict that siRNAs, in addition to being potential therapeutics in their own right, will become increasingly important tools with which to model potential drug effects in vivo, especially if they can be delivered systemically.
Perhaps related to these considerations, mice engineered to have sustained, high-level, hepatic HIF activation develop fatty liver (steatosis) associated with impaired fatty acid oxidation.26,46-50 We have not yet observed such changes in mice treated with EglN siRNA, suggesting that submaximal hepatic HIF activation, especially if episodic, will not translate into significant liver toxicity. Nonetheless, it would be prudent to carefully monitor liver function in patients receiving either organic small-molecule– or siRNA-based EglN inhibitors.
There is an Inside Blood commentary on this article in this issue.
The online version of this article contains a data supplement.
Presented in part at the American Society of Hematology 53rd Annual Meeting and Exposition, December 12, 2011, San Diego, CA.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Julia Hettinger, Chris Zurenko, Justin Aubin, Tim Racie, Will Cantley, Lauren Speciner, and Brian Bettencourt for technical assistance; Dinah Sah, Muthiah Manoharan, and John Maraganore for helpful discussions; and David Nathan and Frank Bunn for critical reading of the manuscript.
These studies were supported by the National Institutes of Health and the Howard Hughes Medical Institute. W.G.K. is a Doris Duke Distinguished Clinical Scientist.
National Institutes of Health
Authorship
Contribution: W.Q., R.L.B., and J.M. designed and performed the experiments and analyzed the data; J.W. and A.Y.C. performed the experiments; E.B., S.K., and A.A. provided critical reagents; K.F. and V.K. designed the experiments and analyzed the data; W.G.K. developed the project, designed the experiments, and analyzed the data; and W.Q., J.M., and W.G.K. wrote the manuscript.
Conflict-of-interest disclosure: W.G.K. owns equity in and is a paid consultant for Fibrogen Inc, which is developing EglN inhibitors, and is a coinventor on a patent related to clinical use of EglN inhibitors that was licensed to Fibrogen Inc. W.Q., J.W., A.Y.C., E.B., S.K., A.A., K.F., and V.K. are employees and stockholders of Alnylam Inc, which is developing RNAi therapeutics. R.L.B. is supported in part by Alnylam Inc. The remaining authors declare no competing financial interests.
Correspondence: William G. Kaelin Jr, Howard Hughes Medical Institute, Brigham and Women's Hospital and Dana-Farber Cancer Institute, 450 Brookline Ave, Boston, MA 02215; e-mail: william_kaelin@dfci.harvard.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal