Abstract
The Wilms tumor protein (WT-1) is widely recognized as a tumor antigen that is expressed differentially by several malignancies. However, WT-1 peptides known to induce tumoricidal T cells are few. In the present study, we evaluated T-cell responses of 56 healthy donors to in vitro sensitization with autologous APCs loaded with a pool of overlapping 15-mer peptides spanning the sequence of WT-1. Thereafter, we mapped the WT-1 peptides eliciting responses in each individual, defined the immunogenic peptides, and identified their presenting HLA alleles. We report 41 previously unreported epitopes of WT-1: 5 presented by class II and 36 by class I alleles, including 10 that could be presented by more than 1 class I allele. IFNγ+ T cells responding to 98% of the class I and 60% of the class II epitopes exhibited HLA-restricted cytotoxicity against peptide-loaded targets. T cells specific for 36 WT-1 peptides were evaluable for leukemocidal activity, of which 27 (75%) lysed WT-1+ leukemic targets sharing their restricting HLA allele. Each epitope identified induced T-cell responses in most donors sharing the epitopes' presenting allele; these responses often exceeded responses to flanking peptides predicted to be more immunogenic. This series of immunogenic epitopes of WT-1 should prove useful for immunotherapies targeting WT-1+ malignancies.
Introduction
After the initial demonstration that infusion of unselected transplantation donor-derived lymphocytes into allogeneic BM transplantation recipients could induce durable complete remissions of chronic myeloid leukemia relapses or EBV+ lymphomas emerging after transplantation,1,2 several groups have been exploring adoptive T-cell therapies for the treatment of chemotherapy-resistant malignancies. Recently, Dudley et al have reported prolonged regressions of melanomas after infusions of in vitro expanded autologous tumor-infiltrating lymphocytes of undefined specificity.3,4 Adoptive therapies using autologous T cells specifically sensitized in vitro against antigens differentially expressed by tumor cells5,6 or genetically modified to express tumor-specific T-cell receptors7,8 or ScFv-based chimeric antigen receptors9–11 have also induced significant responses in a minority of cases. However, these responses have usually been short-lived.
One limitation to the development of effective T-cell immunotherapies is the small number of defined, immunogenic antigens that are both differentially expressed by clonogenic tumor cells and essential to their survival. Recently, antigens with these characteristics have been identified.12–14 One of these, the Wilms tumor protein (WT-1), is a zinc-finger protein encoded by a gene comprising 10 exons located at 11p13.15 Because of extensive RNA splicing, the WT-1 protein is expressed in several isoforms, the balance of which determines its function as a regulator of transcription.16 WT-1 is essential to embryonic development of the urogenital system.17 Postnatally, WT-1 expression in healthy tissues is limited to the ovary, testis, podocytes of the kidney and the mesothelial linings of the peritoneum and pleura.18 WT-1 is also expressed at low levels in hematopoietic progenitor cells, where it normally acts to induce quiescence of CD34+ Lin− cells and promote differentiation of precursors at later stages of development.19,20 In contrast, WT-1 is highly expressed in several solid tumors,21 and in up to 70% of acute myeloid leukemias affecting children and adults; high levels of WT-1 expression are associated with poor prognosis.22–24 WT-1 is also aberrantly expressed in chronic myeloid leukemia25 and in advanced forms of myelodysplasia.26 In leukemic blasts, the balance of WT-1 isoforms expressed appears to promote proliferation and resistance to apoptosis.27 Inhibition of WT-1 expression by ShRNA reverses these effects, thereby eliminating leukemic cells with clonogenic potential.28
Several studies have demonstrated that T cells specific for certain peptides of WT-1 that are expected to be immunogenic based on their predicted binding to specific HLA alleles can be generated from healthy donors and some tumor-bearing patients.29–32 Furthermore, these T cells can lyse WT-1+ tumor cells in vitro29,30,32 and inhibit their growth in vivo in immunodeficient mice bearing human WT-1+ tumor xenografts.32,33 Those peptides found to be immunogenic have subsequently been incorporated into tumor vaccines currently in clinical trials.34–36 However, whereas some of these peptides have consistently elicited T-cell responses in vivo, the antitumor activity of T cells responding to certain of these peptides has been limited. Furthermore, because predictive binding algorithms have been developed for only a limited number of HLA alleles, peptides selected for evaluation have been largely restricted to epitopes predicted to be presented by HLA-A0201, A2402, and DRB10401.
In the present study, we examined the responses of 56 healthy donors to a pool of overlapping pentadecapeptides (15-mers) spanning the sequence of the WT-1 protein. Thereafter, we identified and mapped 41 previously unreported epitopes of WT-1 eliciting T-cell responses, and identified the class I and/or class II HLA allele(s) presenting each peptide. In addition, we have assessed their cytotoxic activity against leukemic cell blasts expressing WT-1 and the peptide's presenting HLA allele and compared the immunogenicity and cytotoxicity of T cells generated against these peptides with that of WT-1 peptides previously predicted to be immunogenic in man.
Methods
WT-1 peptides
The sequence of the WT-1 protein published by Gessler et al,37 which comprises 575 amino acids and includes the first 126 amino acids in the N-terminus missing in the (exon 5+, KTS+) isoform of WT-1,16 was used to design the peptide sequences (see Figure 2A). A total of 141 pentadecapeptides spanning this sequence, each overlapping the next by 11 amino acids, were synthesized by Invitrogen to specifications of validated sequence, 95% purity, sterility, and absence of endotoxin. These one hundred forty-one 15-mers were mixed in equal amounts to form a total pool of peptides, in which each peptide was at a concentration of 0.35 μg/mL. This pool was used for the T-cell sensitization. To identify peptides eliciting responses, subpools containing 12 pentadecapeptides (4.17 μg/mL per peptide) were established to form a mapping matrix in which each peptide was included in only 2 overlapping subpools (Figure 2B).
Generation of WT-1–specific T cells
Peripheral blood was obtained from 56 consenting healthy donors according to protocols approved by the institutional review board of Memorial Sloan-Kettering Cancer Center (New York, NY).
Cytokine-activated monocytes (CAMs) were used as APCs and generated as described previously.32 Briefly, peripheral blood monocytes were separated by adherence on plastic and cultured in RPMI 1640 medium containing 1% autologous serum. GM-CSF (Berlex) and IL-4 (R&D Systems) were added to final concentrations of 2000 and 1000 U/mL, respectively, on days 0, 2, and 4. On day 5, these cells were also treated with TNFα (10 ng/mL), IL-6 (1000 IU/mL), IL-1β (400 IU/mL), and PGE2 (25mM−3; R&D Systems) together with GM-CSF and IL4 at the same doses. CAMs harvested on day 7 of culture expressed CD83, CD80, CD86, and HLA class I and II alleles, as determined by FACS analysis.
EBV-transformed B-lymphoblastoid cell lines (EBV-BLCLs) were also used as WT-1 peptide loaded and control APCs or as targets, as specified in the experiments. They were generated by infection of PBMCs with EBV strain B95.8,38,39 as described previously. EBV-BLCLs were cultured in RPMI 1640 medium (Gemini) with 10% FCS (Gemini) in the presence of acyclovir.
Sensitization and propagation of WT-1 specific T cells.
To generate WT-1–specific cytotoxic T lymphocytes (CTLs), PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation. Monocytes were depleted by adherence on plastic and natural killer cells by absorption to immunomagnetic CD56-precoated microbeads (Miltenyi Biotec).32 Enriched T-cell fractions were stimulated at a 20:1 responder: stimulator ratio with autologous CAMs or EBV-BLCLs that had been preloaded for 3 hours with the total pool of the WT-1 pentadecapeptides in serum-free medium and irradiated to 3000 cGy. T cells were cultured in Yssel medium supplemented with 5% AB human serum (YH5; Gemini), restimulated weekly with autologous WT-1 total pool-loaded CAMs or EBV-BLCLs, and fed with IL-2 (Collaborative Biomedical Products) every 2-3 days at 10-50 U/mL.
Leukemic cells.
Twenty-four primary leukemic cells and 1 leukemic cell line were characterized for their expression of WT-1 by intracellular FACS staining using murine antihuman WT-1 mAbs (Neomarkers) as described previously.32,38 The WT-1+ leukemias included blast cells from 11 primary AMLs, 3 primary ALLs, and 1 B-cell precursor ALL cell line. Ten WT-1− leukemias were used as controls and included 3 B-cell precursor ALLs and 7 AMLs.
All EBV-BLCLs and leukemia cells were typed for HLA-A, HLA-B, HLA-C, HLA-DR, and HLA-DQ alleles at high resolution using standard techniques.
Assessment of T-cell response
IFNγ production by WT-1–specific T cells.
The proportion and phenotype (CD4 and CD8) of T cells generating IFNγ in response to secondary stimulation with the WT-1 total pool, WT-1 subpools, or single WT-1 15-mer or 9-mer WT-1 peptides loaded on autologous PBMCs were measured by FACS analysis of T cells containing intracellular IFNγ, as described previously.38,40
Mapping of immunogenic epitopes.
Aliquots of the T cells stimulated with the WT-1 total pool for 35-42 days were washed and restimulated overnight with autologous PBMCs loaded with one of each of the subpools of WT-1 pentadecapeptides. T-cell responses to each subpool were quantitated by FACS analysis of T cells bearing intracellular IFNγ as described previously.41 The mapping grid (Figure 2B) was then used to identify specific WT-1 15-mers eliciting T-cell responses. These 15-mers and 9-mer or 11-mer sequences within the 15-mers were then analyzed as secondary single-peptide stimulators to confirm their immunogenicity and to define the immunogenic epitope(s) within the 15-mer eliciting responses.
Cytotoxic activity.
The WT-1–specific and HLA-restricted cytotoxic activity of sensitized T cells was measured in standard 51Cr-release assays against a panel of HLA-matched and HLA-mismatched CAM targets either unmodified or loaded with the total pool, the identified 15-mer, or the 9-mer or 11-mer epitope of WT-1 eliciting T-cell responses, as described previously.32 In addition, the restricting HLA allele presenting each immunogenic epitope was identified by measuring the cytotoxicity of the sensitized T cells against a panel of allogeneic CAMs preloaded with the peptide, each sharing a single HLA allele expressed on the responding WT-1–specific T cells, as described previously.41 The cytotoxic activity of the WT-1 epitope–specific CTLs against WT-1− and WT-1+ leukemia cell lines or primary leukemic cells expressing the restricting HLA alleles was also assessed in this cytotoxicity assay, as described previously.32
Immunogenicity of the identified immunodominant WT-1–derived epitopes
To estimate the immunogenicity of identified WT-1 peptide epitopes in different subjects, enriched T cells separated from PBMCs of healthy donors expressing 1 of a series of prevalent HLA alleles (ie, HLA-A0201, HLA-A0301, HLA-A2402, or HLA-B0702), which we had identified as a presenter of a newly identified WT-1 epitope, were sensitized in vitro with artificial APCs (AAPCs)42 expressing that HLA allele and loaded with the preidentified WT-1 epitope or an irrelevant peptide. The panel of AAPCs included those expressing one of the following single HLA alleles: HLA-A0201, HLA-A0101, HLA-A0301, HLA-A2402, HLA-B0702, or HLA-B0801, which were generated as described previously.42 After 35 days of coculture of T cells with the peptide-loaded AAPCs in the presence of IL-2, CTLs were secondarily stimulated overnight with autologous PBMCs loaded with the sensitizing peptide or an unrelated peptide and tested for their IFNγ response. The responses were registered as positive if the proportion of T cells producing IFNγ in response to secondary stimulation with autologous PBMCs loaded with the stimulating WT-1–derived peptide exceeded the background proportion of IFNγ T cells incubated with PBMCs alone by 2-fold or more.
Results
Responses of healthy donors to the WT-1 total pool of pentadecapeptides
We initially measured the frequencies of WT-1–specific IFNγ+ T cells in the PBMCs of 41 healthy donors. These frequencies ranged between 0.01% and 1.82% and exceeded the background of IFNγ+ T cells detected in T cells stimulated with autologous PBMCs alone in only 10 of 41 subjects (Figure 1A). In vitro sensitization of T cells from 56 healthy donors with autologous CAMs loaded with the total pool of WT-1 pentadecapeptides for periods of 35-42 days resulted in significant expansion of IFNγ+ T cells in 41 of 56 cases (73%; Figure 1A). T cells generated from 38 of 56 donors also exhibited cytotoxic activity against autologous phytohemagglutinin (PHA) blasts loaded with the WT-1 total pool (Figure 1B), including T cells from 38 of the 41 donors who produced IFNγ in response to secondary stimulation with the WT-1 peptide pool.
WT-1–specific responses of CTLs generated from PBMCs of healthy donors (n = 56) by stimulation with autologous APCs loaded with total pool of WT-1–derived pentadecapeptides. (A) production of IFNγ in PBMCs alone (as a background), PBMCs coincubated overnight with the total pool of pentadecapeptides spanning the whole sequence of WT-1 protein (PBMC + WT-1 pool), and pregenerated WT-1–specific T cells coincubated overnight with WT-1 peptide–loaded PBMCs. (B) Cytotoxic activity of the WT-1–specific CTLs generated in vitro by stimulation with WT-1 total pool against WT-1− (autologous PHA-stimulated blasts) and WT-1+ (autologous PHA-stimulated blasts loaded with the total pool of WT-1 pentadecapeptides) targets at a 50:1 effector: stimulator ratio. (C) IFNγ response measured by FACS staining in different responder cell populations (PBMCs, pregenerated CTLs sensitized in vitro with the RMF peptide loaded on autologous CAM and pregenerated CTLs sensitized with the total pool of WT-1 15-mers) after secondary overnight stimulation with autologous PBMCs either unmodified or loaded with one of the following: RMF peptide, dominant epitopes of WT-1 identified by the epitope-mapping approach in the WT-1–total pool sensitized CTLs, or the WT-1 total pool of the 141 pentadecapeptides. (D) Cytotoxic activity of the WT-1–specific T cells generated in vitro by sensitization with autologous CAMs loaded with the RMF 9-mer or with the total pool of the WT-1 15-mers. The cytotoxicity of the T cells was assessed against autologous WT-1− targets (PHA-activated blasts) and the same targets loaded with RMF peptide, the total pool of WT-1 15-mers, or the dominant WT-1 epitope identified for the same T-cell line.
WT-1–specific responses of CTLs generated from PBMCs of healthy donors (n = 56) by stimulation with autologous APCs loaded with total pool of WT-1–derived pentadecapeptides. (A) production of IFNγ in PBMCs alone (as a background), PBMCs coincubated overnight with the total pool of pentadecapeptides spanning the whole sequence of WT-1 protein (PBMC + WT-1 pool), and pregenerated WT-1–specific T cells coincubated overnight with WT-1 peptide–loaded PBMCs. (B) Cytotoxic activity of the WT-1–specific CTLs generated in vitro by stimulation with WT-1 total pool against WT-1− (autologous PHA-stimulated blasts) and WT-1+ (autologous PHA-stimulated blasts loaded with the total pool of WT-1 pentadecapeptides) targets at a 50:1 effector: stimulator ratio. (C) IFNγ response measured by FACS staining in different responder cell populations (PBMCs, pregenerated CTLs sensitized in vitro with the RMF peptide loaded on autologous CAM and pregenerated CTLs sensitized with the total pool of WT-1 15-mers) after secondary overnight stimulation with autologous PBMCs either unmodified or loaded with one of the following: RMF peptide, dominant epitopes of WT-1 identified by the epitope-mapping approach in the WT-1–total pool sensitized CTLs, or the WT-1 total pool of the 141 pentadecapeptides. (D) Cytotoxic activity of the WT-1–specific T cells generated in vitro by sensitization with autologous CAMs loaded with the RMF 9-mer or with the total pool of the WT-1 15-mers. The cytotoxicity of the T cells was assessed against autologous WT-1− targets (PHA-activated blasts) and the same targets loaded with RMF peptide, the total pool of WT-1 15-mers, or the dominant WT-1 epitope identified for the same T-cell line.
We also compared the capacity of one of the previously reported WT-1 epitopes predicted to bind the HLA-A0201 allele, 126-134RMFPNAPYL (RMF),43 and the total pool of WT-1 pentadecapeptides to stimulate WT-1 reactive T cells in HLA-A0201+ healthy donors (n = 14) when loaded on autologous CAMs. Increased frequencies of IFNγ+ T cells initially sensitized with the RMF peptide were detected in 9 of 14 donors, 7 of whom also responded to secondary simulation with the pooled peptides (Figure 1C). In contrast, 12 of 14 CTL lines initially sensitized with the WT-1 peptide pool generated high frequencies of IFNγ+ T cells after secondary stimulation with the WT-1 total pool, including 6 CTL lines that also responded to RMF. We mapped the epitopes of WT-1 recognized by the T cells sensitized with the total pool (vide infra) and identified epitopes other than RMF in 12 of 14 donors. The magnitude of the responses to those epitopes was much higher than to the RMF peptide (Figure 1C). Only 4 of 14 CTL lines initially sensitized with RMF exhibited cytotoxic activity against RMF-loaded autologous PHA blasts; of these, 3 could also lyse autologous PHA blasts loaded with the WT-1 pool (Figure 1D). In contrast, 10 of 14 CTLs sensitized with the pool of WT-1 peptides were cytotoxic against PHA blasts loaded with the WT-1 total pool, including 3 of 14 that lysed RMF peptide–loaded blasts (Figure 1D). Therefore, in a high proportion of HLA-A0201+ donors, stimulation of T cells with the WT-1 total pool more consistently elicited WT-1–specific T-cell responses than stimulation with the single HLA-A0201–binding RMF peptide.
WT-1 CTLs generated by sensitization with the pooled peptides are epitope specific and HLA restricted
The epitopes recognized by T cells sensitized in vitro with the total pool of overlapping WT-1 pentadecapeptides (Figure 2A) were identified by quantitating IFNγ+ T cells responding to the mapping grid of subpools of WT-1 15-mers (Figure 2B). As seen in the representative example provided in Figure 2C, significantly increased numbers of IFNγ+ T cells were selectively generated in response to subpools no. 3 and 19, which share the pentadecapeptide no. 75. The T cells were then stimulated with neighboring 15-mers, each overlapping peptide no. 75 by 11 amino acids. As can be seen, IFNγ+ T cells are selectively generated in response to peptide no. 75 (Figure 2D). The newly identified immunogenic WT-1 epitope is 174-182HSFKHEDPM. Subsequently, we assessed the cytotoxic activity of these T cells against a panel of allogeneic CAMs either unmodified or loaded with this peptide, each sharing 1 HLA allele expressed by the tested CTLs. As shown in Figure 2E, the T cells selectively lysed peptide-loaded autologous targets and targets expressing the HLA-B3501 allele and did not lyse peptide-loaded targets sharing other HLA alleles inherited by the T-cell donor. These T cells also lysed WT-1+ BALL cells coexpressing the HLA-B3501 allele.
Strategy for the generation of the total pool of overlapping pentadecapeptides spanning the whole sequence of the WT-1 protein and epitope mapping. (A) The sequence of the WT-1 protein consisting of 575 amino acids and the principle of 11 amino acid–overlapping pentadecapeptides are illustrated. A total of 141 pentadecapeptides are required to span the entire protein. The sequence of 575 amino acids published by Gessler et al was used.37 This sequence includes an additional 126 amino acids in the N-terminus. To match the sequential numbers of amino acids within the WT-1 sequence used with the longest, most frequently described WT-1 isoform D, we numbered the first 126 amino acids with negative values and used the positive values to number the subsequent 449 amino acids described in the longest isoform D. (B) Mapping grid consisting of 24 subpools each containing up to 12 WT-1–derived pentadecapeptides. Each peptide is uniquely contained within 2 intersecting subpools: for example, peptide 75 is uniquely shared by subpools 3 and 19. (C) IFNγ production by WT-1–sensitized CTLs in response to secondary overnight stimulation with the subpools of WT-1 pentadecapeptides loaded on autologous PBMCs. Dominant responses are observed for the subpools no. 3 and 19, both containing 1 common pentadecapeptide no. 75. (D) IFNγ production by the WT-1 CTLs in response to secondary overnight stimulation with the single pentadecapeptide contained within the subpools eliciting the highest responses as per the analysis determined in panel C confirms that the dominant immunogenic sequence is contained within pentadecapeptide no. 75. (E) HLA restriction of the WT-1–specific T cells responding to peptide no. 75 identified by 51Cr-release assay against a panel of allogeneic CAMs or PHA blasts matching single HLA alleles expressed by the WT-1 CTL donors. These are presented along the x-axis of the graph. The CAMs or PHA blasts used in the assay are unmodified (gray bars) or loaded with the WT-1–dominant epitope (black bars). The WT-1–specific cytotoxic activity of the WT-1 CTLs is restricted by the B3501 HLA allele.
Strategy for the generation of the total pool of overlapping pentadecapeptides spanning the whole sequence of the WT-1 protein and epitope mapping. (A) The sequence of the WT-1 protein consisting of 575 amino acids and the principle of 11 amino acid–overlapping pentadecapeptides are illustrated. A total of 141 pentadecapeptides are required to span the entire protein. The sequence of 575 amino acids published by Gessler et al was used.37 This sequence includes an additional 126 amino acids in the N-terminus. To match the sequential numbers of amino acids within the WT-1 sequence used with the longest, most frequently described WT-1 isoform D, we numbered the first 126 amino acids with negative values and used the positive values to number the subsequent 449 amino acids described in the longest isoform D. (B) Mapping grid consisting of 24 subpools each containing up to 12 WT-1–derived pentadecapeptides. Each peptide is uniquely contained within 2 intersecting subpools: for example, peptide 75 is uniquely shared by subpools 3 and 19. (C) IFNγ production by WT-1–sensitized CTLs in response to secondary overnight stimulation with the subpools of WT-1 pentadecapeptides loaded on autologous PBMCs. Dominant responses are observed for the subpools no. 3 and 19, both containing 1 common pentadecapeptide no. 75. (D) IFNγ production by the WT-1 CTLs in response to secondary overnight stimulation with the single pentadecapeptide contained within the subpools eliciting the highest responses as per the analysis determined in panel C confirms that the dominant immunogenic sequence is contained within pentadecapeptide no. 75. (E) HLA restriction of the WT-1–specific T cells responding to peptide no. 75 identified by 51Cr-release assay against a panel of allogeneic CAMs or PHA blasts matching single HLA alleles expressed by the WT-1 CTL donors. These are presented along the x-axis of the graph. The CAMs or PHA blasts used in the assay are unmodified (gray bars) or loaded with the WT-1–dominant epitope (black bars). The WT-1–specific cytotoxic activity of the WT-1 CTLs is restricted by the B3501 HLA allele.
Mapping of WT-1 peptides eliciting T-cell responses identifies a diversity of immunogenic epitopes presented by different class I and II HLA alleles
We used the same approach to map and ultimately identify WT-1 epitopes eliciting responses by T cells from the other 40 responding healthy donors. Of these donors, 8 (19%) responded exclusively to 1 WT-1 peptide, whereas 18 (43%) responded to 2 peptides and 16 (39%) to 3 peptides. In cultures eliciting responses to more than 1 WT-1 peptide, the patterns of IFNγ+ T-cell responses to the subpools were sufficiently distinctive to permit initial segregation of potentially immunogenic peptides. Each candidate peptide was then evaluated individually to ascertain the specific peptide inducing a T-cell response.
The immunogenic peptides of WT-1 that we have identified and their presenting HLA alleles are listed in Table 1. Of the 42 WT-1 peptides eliciting T-cell responses, 41 are newly identified; only one, the 126-134RMFPNAPYL nonamer presented by HLA-A0201, has been described previously and shown to be immunogenic when presented by this allele.43 Peptide 91, 235-249CMTWNQMNLGATLKG, contains an epitope that elicited CD4+ T-cell responses restricted by HLA-DRB1 0402, but also contains the 235-243CMT nonamer known to be presented by HLA-A0201 and HLA-A2402.29 For 26 of the peptides presented by class I HLA alleles, we identified a single presenting HLA allele in the initially studied donor. However, when we examined the HLA restrictions of T cells responding to these peptides in different donors, we found that 10 of these peptides could elicit T-cell responses when presented by 2 or 3 different class I HLA alleles. One sequence, the 238-246WNQMNLGAT peptide, elicited strong IFNγ+ CD8+ T-cell responses when presented in different donors by any 1 of 4 distinct HLA class I alleles.
WT-1−derived immunogenic epitopes identified by mapping the IFNγ responses of T cells after sensitization with the pool of overlapping pentadecapeptides spanning the whole sequence of the WT-1 protein
| 15-mer no. containing the dominant epitope . | Sequence identified . | Presenting HLA allele . | IFNγ response of T cells, % IFNγ+ cells . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs: . | ||||
|---|---|---|---|---|---|---|---|---|
| No WT-1 peptide . | WT-1 peptide loaded . | WT-1− autologous APCs . | WT-1 peptide–loaded autologous APCs . | WT-1− leukemia cells . | WT-1+ leukemia cells* . | |||
| 1 | (−125)-(−117)RQRPHPGAL | B0702 | 0.9 | 11.3 | 0 | 27 | 1 | 67 |
| 2 | (−119)-(−111)GALRNPTAC | B0702 | 0.5 | 14.0 | 0 | 30 | 1 | 60 |
| 4 | (−110)-(−102)PLPHFPPSL | A0201 | 0.98 | 5.75 | 0 | 30 | 2 | 22 |
| 5 | (−107)-(−99)HFPPSLPPT | A3101 | 0.73 | 4.82 | 0 | 42 | ND | ND |
| 7 | (−99)-(−91)THSPTHPPR | B4001 | 1.5 | 12.8 | 0 | 45 | 3 | 65 |
| A0201 | 0.4 | 5 | 2 | 50 | 0 | 38 | ||
| 13 | (−75)-(67)AILDFLLLQ | A0201 | 0.61 | 5.07 | 0 | 18 | 3 | 19 |
| 20 | (−47)-(39)PGCLQQPEQ | A0201 | 0.2 | 3.67 | 6 | 54 | 5 | 19 |
| B4701 | 0.5 | 4.6 | 6 | 54 | ND | ND | ||
| (−47)-(−37)PGCLQQPEQQG | DRB10101 | 0.33 | 3.1 | 6 | 54 | ND | ND | |
| 24-25 | (−27)-(−19)KLGAAEASA | A0201 | 1.05 | 4.48 | 3 | 41 | 10 | 37 |
| 29-30 | (−8)-(1)ASGSEPQQM | B3501 | 0.07 | 1.0 | 5 | 73 | 5 | 39 |
| 33 | 6-15RDLNALLPAV | A0201 | 1.1 | 11.0 | 2 | 51 | 0 | 9 |
| B5701 | 0.19 | 1.24 | 3 | 44 | ND | ND | ||
| 37 | 22-31GGCALPVSGA | A0201 | 0.07 | 0.9 | 8 | 32 | 3 | 47 |
| 39 | 30-38GAAQWAPVL | B3901 | 0.1 | 1.3 | 2 | 31 | ND | ND |
| 41 | 38-46LDFAPPGAS | A0201 | 0.2 | 4.18 | 0 | 73 | 0 | 40 |
| 38-48LDFAPPGASAY | DRB10402 | 0.2 | 1.41 | 0 | 73 | 0 | 40 | |
| 43 | 46-54SAYGSLGGP | A0201 | 1.2 | 6.46 | 2 | 51 | 0 | 0 |
| B4001 | 1.09 | 6.84 | 2 | 41 | 3 | 68 | ||
| 46 | 58-66PAPPPPPPP | A0201 | 1.15 | 6.69 | 2 | 40 | 0 | 0 |
| 58 | 106-114ACRYGPFGP | B4402 | 0.92 | 5.65 | 8 | 46 | ND | ND |
| 62 | 122-130SGQARMFPN | B3503 | 0.78 | 2.0 | 0 | 84 | ND | ND |
| C0401 | 0.78 | 2.0 | 0 | 84 | ND | ND | ||
| 62-63 | 126-134RMFPNAPYL† | A0201 | 0.52 | 2.17 | 3 | 41 | 2 | 25 |
| 65-66 | 135-143PSCLESQPA | B3501 | 0.07 | 0.61 | 0 | 35 | ND | ND |
| 68 | 146-154NQGYSTVTF | A0101 | 0.92 | 4.0 | 2 | 19 | ND | ND |
| 73 | 166-174HHAAQFPNH | B3801 | 0.81 | 3.14 | 0 | 26 | ND | ND |
| 74-75 | 174-182HSFKHEDPM | B3501 | 1.3 | 18.0 | 0 | 50 | 5 | 45 |
| 82 | 202-210CHTPTDSCT | B4402 | 1.02 | 3.77 | 8 | 37 | ND | ND |
| 83-84 | 209-217CTGSQALLL | A0101 | 0.03 | 0.29 | 0 | 21 | 3 | 33 |
| 83 | 206-214TDSCTGSQA | B3802 | 0.71 | 4.02 | 0 | 88 | ND | ND |
| B4402 | 1.01 | 4.2 | 1 | 36 | 1 | 56 | ||
| 86 | 218-226RTPYSSDNL | B3503 | 0.84 | 3.0 | 0 | 84 | 4 | 48 |
| C0401 | 0.84 | 3.0 | 0 | 84 | 4 | 48 | ||
| 87 | 225-233NLYQMTSQLE | A0201 | 0.13 | 0.9 | 3 | 87 | 0 | 0 |
| 91 | 238-246WNQMNLGAT | A0201 | 1.34 | 8.0 | 0 | 18 | 1 | 19 |
| C1701 | 2.1 | 12.0 | 0 | 10 | 1 | 16 | ||
| A0101 | 2.1 | 7.31 | 0 | 26 | ND | ND | ||
| B3508 | 1.23 | 5.0 | 0 | 18 | 4 | 19 | ||
| 91-92 | 239-248NQMNLGATL | A2402 | 0.02 | 0.14 | 4 | 9 | 1 | 17 |
| 91 | 238-248WNQMNLGATLK | DRB11104 | 0.59 | 6.0 | 0 | 8 | 0 | 0 |
| 235-249CMTWNQMNLGATLKG | DRB10402 | 0.07 | 0.53 | 4 | 16 | 1 | 17 | |
| 92 | 242-250NLGATLKGV | A0101 | 0.32 | 1.83 | 2 | 19 | ND | ND |
| A0201 | 0.06 | 0.75 | 1 | 18 | 2 | 19 | ||
| 92-93 | 243-252LGATLKGVAA | A0203 | 0.54 | 2.1 | 0 | 35 | ND | ND |
| 93 | 246-253TLKGVAAGS | A6901 | 0.09 | 1.85 | 4 | 80 | ND | ND |
| 99-100 | 269-278GYESDNHTT | A0101 | 0.12 | 2.43 | 0 | 27 | 0 | 33 |
| B3501 | 0.1 | 0.61 | 0 | 35 | ND | ND | ||
| 112-113 | 323-332FMCAYPGCNK | B3501 | 1.3 | 18.0 | 0 | 70 | 5 | 45 |
| 320-334KRPFMCAYPGC | DRB10401 | 0.91 | 3.48 | 9 | 5 | 5 | 5 | |
| 129 | 390-398RKFSRSDHL | A0201 | 1.08 | 5.81 | 3 | 40 | ND | ND |
| 131 | 398-406LKTHTTRTHT | A0201 | 1.56 | 14.0 | 0 | 38 | ND | ND |
| 141 | 436-445NMHQRNHTKL | A0201 | 1.78 | 6.69 | 2 | 40 | 0 | 0 |
| B4001 | 2.1 | 7.71 | 0 | 31 | 3 | 72 | ||
| A2402 | 0.61 | 2.79 | 19 | 47 | 0 | 0 | ||
| 15-mer no. containing the dominant epitope . | Sequence identified . | Presenting HLA allele . | IFNγ response of T cells, % IFNγ+ cells . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs: . | ||||
|---|---|---|---|---|---|---|---|---|
| No WT-1 peptide . | WT-1 peptide loaded . | WT-1− autologous APCs . | WT-1 peptide–loaded autologous APCs . | WT-1− leukemia cells . | WT-1+ leukemia cells* . | |||
| 1 | (−125)-(−117)RQRPHPGAL | B0702 | 0.9 | 11.3 | 0 | 27 | 1 | 67 |
| 2 | (−119)-(−111)GALRNPTAC | B0702 | 0.5 | 14.0 | 0 | 30 | 1 | 60 |
| 4 | (−110)-(−102)PLPHFPPSL | A0201 | 0.98 | 5.75 | 0 | 30 | 2 | 22 |
| 5 | (−107)-(−99)HFPPSLPPT | A3101 | 0.73 | 4.82 | 0 | 42 | ND | ND |
| 7 | (−99)-(−91)THSPTHPPR | B4001 | 1.5 | 12.8 | 0 | 45 | 3 | 65 |
| A0201 | 0.4 | 5 | 2 | 50 | 0 | 38 | ||
| 13 | (−75)-(67)AILDFLLLQ | A0201 | 0.61 | 5.07 | 0 | 18 | 3 | 19 |
| 20 | (−47)-(39)PGCLQQPEQ | A0201 | 0.2 | 3.67 | 6 | 54 | 5 | 19 |
| B4701 | 0.5 | 4.6 | 6 | 54 | ND | ND | ||
| (−47)-(−37)PGCLQQPEQQG | DRB10101 | 0.33 | 3.1 | 6 | 54 | ND | ND | |
| 24-25 | (−27)-(−19)KLGAAEASA | A0201 | 1.05 | 4.48 | 3 | 41 | 10 | 37 |
| 29-30 | (−8)-(1)ASGSEPQQM | B3501 | 0.07 | 1.0 | 5 | 73 | 5 | 39 |
| 33 | 6-15RDLNALLPAV | A0201 | 1.1 | 11.0 | 2 | 51 | 0 | 9 |
| B5701 | 0.19 | 1.24 | 3 | 44 | ND | ND | ||
| 37 | 22-31GGCALPVSGA | A0201 | 0.07 | 0.9 | 8 | 32 | 3 | 47 |
| 39 | 30-38GAAQWAPVL | B3901 | 0.1 | 1.3 | 2 | 31 | ND | ND |
| 41 | 38-46LDFAPPGAS | A0201 | 0.2 | 4.18 | 0 | 73 | 0 | 40 |
| 38-48LDFAPPGASAY | DRB10402 | 0.2 | 1.41 | 0 | 73 | 0 | 40 | |
| 43 | 46-54SAYGSLGGP | A0201 | 1.2 | 6.46 | 2 | 51 | 0 | 0 |
| B4001 | 1.09 | 6.84 | 2 | 41 | 3 | 68 | ||
| 46 | 58-66PAPPPPPPP | A0201 | 1.15 | 6.69 | 2 | 40 | 0 | 0 |
| 58 | 106-114ACRYGPFGP | B4402 | 0.92 | 5.65 | 8 | 46 | ND | ND |
| 62 | 122-130SGQARMFPN | B3503 | 0.78 | 2.0 | 0 | 84 | ND | ND |
| C0401 | 0.78 | 2.0 | 0 | 84 | ND | ND | ||
| 62-63 | 126-134RMFPNAPYL† | A0201 | 0.52 | 2.17 | 3 | 41 | 2 | 25 |
| 65-66 | 135-143PSCLESQPA | B3501 | 0.07 | 0.61 | 0 | 35 | ND | ND |
| 68 | 146-154NQGYSTVTF | A0101 | 0.92 | 4.0 | 2 | 19 | ND | ND |
| 73 | 166-174HHAAQFPNH | B3801 | 0.81 | 3.14 | 0 | 26 | ND | ND |
| 74-75 | 174-182HSFKHEDPM | B3501 | 1.3 | 18.0 | 0 | 50 | 5 | 45 |
| 82 | 202-210CHTPTDSCT | B4402 | 1.02 | 3.77 | 8 | 37 | ND | ND |
| 83-84 | 209-217CTGSQALLL | A0101 | 0.03 | 0.29 | 0 | 21 | 3 | 33 |
| 83 | 206-214TDSCTGSQA | B3802 | 0.71 | 4.02 | 0 | 88 | ND | ND |
| B4402 | 1.01 | 4.2 | 1 | 36 | 1 | 56 | ||
| 86 | 218-226RTPYSSDNL | B3503 | 0.84 | 3.0 | 0 | 84 | 4 | 48 |
| C0401 | 0.84 | 3.0 | 0 | 84 | 4 | 48 | ||
| 87 | 225-233NLYQMTSQLE | A0201 | 0.13 | 0.9 | 3 | 87 | 0 | 0 |
| 91 | 238-246WNQMNLGAT | A0201 | 1.34 | 8.0 | 0 | 18 | 1 | 19 |
| C1701 | 2.1 | 12.0 | 0 | 10 | 1 | 16 | ||
| A0101 | 2.1 | 7.31 | 0 | 26 | ND | ND | ||
| B3508 | 1.23 | 5.0 | 0 | 18 | 4 | 19 | ||
| 91-92 | 239-248NQMNLGATL | A2402 | 0.02 | 0.14 | 4 | 9 | 1 | 17 |
| 91 | 238-248WNQMNLGATLK | DRB11104 | 0.59 | 6.0 | 0 | 8 | 0 | 0 |
| 235-249CMTWNQMNLGATLKG | DRB10402 | 0.07 | 0.53 | 4 | 16 | 1 | 17 | |
| 92 | 242-250NLGATLKGV | A0101 | 0.32 | 1.83 | 2 | 19 | ND | ND |
| A0201 | 0.06 | 0.75 | 1 | 18 | 2 | 19 | ||
| 92-93 | 243-252LGATLKGVAA | A0203 | 0.54 | 2.1 | 0 | 35 | ND | ND |
| 93 | 246-253TLKGVAAGS | A6901 | 0.09 | 1.85 | 4 | 80 | ND | ND |
| 99-100 | 269-278GYESDNHTT | A0101 | 0.12 | 2.43 | 0 | 27 | 0 | 33 |
| B3501 | 0.1 | 0.61 | 0 | 35 | ND | ND | ||
| 112-113 | 323-332FMCAYPGCNK | B3501 | 1.3 | 18.0 | 0 | 70 | 5 | 45 |
| 320-334KRPFMCAYPGC | DRB10401 | 0.91 | 3.48 | 9 | 5 | 5 | 5 | |
| 129 | 390-398RKFSRSDHL | A0201 | 1.08 | 5.81 | 3 | 40 | ND | ND |
| 131 | 398-406LKTHTTRTHT | A0201 | 1.56 | 14.0 | 0 | 38 | ND | ND |
| 141 | 436-445NMHQRNHTKL | A0201 | 1.78 | 6.69 | 2 | 40 | 0 | 0 |
| B4001 | 2.1 | 7.71 | 0 | 31 | 3 | 72 | ||
| A2402 | 0.61 | 2.79 | 19 | 47 | 0 | 0 | ||
Epitopes are identified by position on the WT-1 protein. T cells are characterized as to their production of IFNγ and their cytotoxic activity against peptide loaded APCs and unloaded WT-1+ and WT-1− leukemic cells.
ND indicates not determined.
Leukemia targets were derived from either immortalized leukemia cell lines or primary leukemia cells obtained from patients with WT-1+ leukemia.
The epitope previously predicted by the computer algorithm or described in the literature.
Using this epitope-mapping strategy, we also identified 5 new 11-mer peptides that stimulated CD4+ T-cell responses restricted by HLA class II alleles. The CD4+ T cells generated in response to each of these epitopes expressed high levels of IFNγ+ T cells. The CD4+ T cells responding to 3 of these 5 peptide epitopes also exhibited specific cytotoxic activity against peptide-loaded PHA blasts and unmodified WT-1+ leukemic blasts selectively sharing the restricting class II HLA allele.
In 4 of the 56 donors tested, epitope mapping identified specific 15-mers eliciting both CD4+ and CD8+ T-cell responses (15-mer peptides 20, 41, 91, and 112). Fine mapping of the sequences eliciting these responses identified 4 11-mers that stimulated HLA class II–restricted CD4+ T-cell responses that also contained within their sequences 9-mers that elicited HLA class I–restricted CD8+ T-cell responses. A representative example of one of these dual stimulating peptides is presented in Figure 3. In this case, peptide 41 was found to elicit both CD4+ and CD8+ IFNγ+ T-cell responses (Figure 3A). Fine mapping of the 11-mers within peptide 41 eliciting the CD4+ IFNγ+ T-cell response (Figure 3A) suggested the 38-48LDFAPPGASAY peptide as the most immunogenic sequence inducing both CD4+ and CD8+ IFNγ+ T-cell responses. Strikingly, peptide 41–sensitized T cells lysed PHA blasts sensitized with either the 9–amino acid sequence (38-46LDFAPPGAS) or the 11–amino acid sequence (38-48LDFAPPGASAY), but did not lyse PHA blasts loaded with the 36-46PVLDFAPPGAS or 37-47VLDFAPPGASA 11-mers. Subsequent examination of the HLA restriction of T cells in the culture (Figure 3D) revealed that the class II HLA–restricted T cells were selectively cytotoxic against targets sharing the alleles DRB1 0402 and DQB1 0302 only when loaded with the LDF 11-mer, whereas the T cells restricted by HLA-A0201 were able to lyse targets loaded with either the 11-mer or the 9-mer LDF peptide. In this case, we were not able to ascertain whether DRB10402 or DQB10302 was the restricting class II HLA allele because we did not have cells in the panel expressing one without the other.
HLA class I– and class II–restricted, WT-1–specific T cells respond to the same immunodominant peptide 15-mer derived from WT-1 protein in the WT-1 CTL sensitized with the WT-1 total pool of overlapping 15-mers loaded on autologous CAMs. (A) Production of IFNγ by the CD8+ and CD4+ WT-1–specific T cells in response to secondary overnight stimulation with the same dominant WT-1–derived 15-mer no. 41. (B) Identification of the immunogenic sequence of amino acids within pentadecapeptide no. 41 by IFNγ production after secondary overnight stimulation with autologous PBMCs loaded with a panel of 9-mers either unique for the peptide no. 41 (LDFAAPGAS [LDF]) or contained within the neighboring overlapping 15-mer no. 40 (PVLDFAPPG [PVL] or VLDFAPPGA [VLD]) and no. 42 (DFAPPGASA [DFA]). Only the 9-mer uniquely presented within the 15-mer no. 41, LDF, elicited an IFNγ response. (C) Peptide-specific cytotoxic activity of WT-1 CTLs against the panel of 9-mers and 11-mers contained within peptide no. 41 and loaded on autologous PHA-stimulated blasts is observed against both the 11-mer LDF and 9-mer LDF contained within the 11-mer LDF, as determined in a standard 51Cr-release assay at a 25:1 effector: stimulator ratio. (D) HLA restriction of the cytotoxic activity of the WT-1 CTLs. T cells restricted by HLA-A0201 lyse targets loaded with either the 11-mer or the 9-mer, whereas those restricted by HLA DRB10402 only lysed targets loaded with the 11-mer.
HLA class I– and class II–restricted, WT-1–specific T cells respond to the same immunodominant peptide 15-mer derived from WT-1 protein in the WT-1 CTL sensitized with the WT-1 total pool of overlapping 15-mers loaded on autologous CAMs. (A) Production of IFNγ by the CD8+ and CD4+ WT-1–specific T cells in response to secondary overnight stimulation with the same dominant WT-1–derived 15-mer no. 41. (B) Identification of the immunogenic sequence of amino acids within pentadecapeptide no. 41 by IFNγ production after secondary overnight stimulation with autologous PBMCs loaded with a panel of 9-mers either unique for the peptide no. 41 (LDFAAPGAS [LDF]) or contained within the neighboring overlapping 15-mer no. 40 (PVLDFAPPG [PVL] or VLDFAPPGA [VLD]) and no. 42 (DFAPPGASA [DFA]). Only the 9-mer uniquely presented within the 15-mer no. 41, LDF, elicited an IFNγ response. (C) Peptide-specific cytotoxic activity of WT-1 CTLs against the panel of 9-mers and 11-mers contained within peptide no. 41 and loaded on autologous PHA-stimulated blasts is observed against both the 11-mer LDF and 9-mer LDF contained within the 11-mer LDF, as determined in a standard 51Cr-release assay at a 25:1 effector: stimulator ratio. (D) HLA restriction of the cytotoxic activity of the WT-1 CTLs. T cells restricted by HLA-A0201 lyse targets loaded with either the 11-mer or the 9-mer, whereas those restricted by HLA DRB10402 only lysed targets loaded with the 11-mer.
T cells generated against newly identified WT-1 epitopes exhibit cytotoxic activity against WT-1+ leukemias
Once we established the WT-1 peptide specificity and HLA restrictions of the IFNγ+ T cells responding to the pool of WT-1 peptides, we examined their cytotoxic activity against: (1) unmodified and peptide-loaded autologous PHA blasts and (2) a series of allogeneic PHA blasts loaded with the identified peptides and primary acute leukemic cell blasts expressing WT-1 protein that coexpressed the WT-1 specific T cells' restricting HLA allele. For the latter tests, WT-1+ leukemic cells not expressing the restricting allele and WT-1− cells sharing the restricting allele served as controls. Results are summarized in Tables 1 and 2.
WT-1–derived immunogenic epitopes identified by mapping the IFNγ responses of T cells after sensitization with the pool of overlapping pentadecapeptides spanning the whole sequence of the WT-1 protein
| Presenting HLA allele . | Sequence identified . | Prediction algorithm . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs: . | ||||
|---|---|---|---|---|---|---|---|
| Binding index . | Disassociation time . | WT-1− allo-APCs with restricting HLA allele . | WT-1+ allo-APCs with restricting HLA allele loaded with WT-1 peptide . | WT-1− leukemia cells . | WT-1+ leukemia cells . | ||
| A0101 | 146-154NQGYSTVTF | 3 | 0.001 | 4 | 15 | ND | ND |
| 209-217CTGSQALLL | 12 | 0.125 | 0 | 26 | 3 | 33 | |
| 238-246WNQMNLGAT | 2 | 0 | 3 | 19 | ND | ND | |
| 242-250NLGATLKGV | 3 | 0.01 | 1 | 17 | ND | ND | |
| 269-278GYESDNHTT | 15 | 1.5 | 0 | 26 | 0 | 33 | |
| 323-332FMCAYPGCNK* | 0 | 0.1 | 2 | 0 | 5 | 0 | |
| A0201 | (−110)-(−102)PLPHFPPSL | 21 | 2 | 1 | 24 | 2 | 22 |
| (−99)-(−91)THSPTHPPR | 3 | 0 | 1 | 21 | 0 | 38 | |
| (−75)-(−67)AILDFLLLQ | 19 | 0.272 | 3 | 17 | 3 | 19 | |
| (−47)-(−39)PGCLQQPEQ | 0 | 0 | 7 | 27 | 5 | 19 | |
| (−27)-(−19)KLGAAEASA | 19 | 17 | 2 | 22 | 10 | 37 | |
| 6-15RDLNALLPAV | 18 | 0.2 | 4 | 31 | 0 | 9 | |
| 22-31GGCALPVSGA | 13 | 0.003 | 3 | 25 | 3 | 47 | |
| 38-46LDFAPPGAS | 11 | 0 | 1 | 62 | 0 | 40 | |
| 46-54SAYGSLGGP* | 14 | 0 | 5 | 31 | 0 | 0 | |
| 58-66PAPPPPPPP* | 5 | 0 | 1 | 18 | 0 | 0 | |
| 126-134RMFPNAPYL† | 22 | 313 | 1 | 52 | 2 | 25 | |
| 225-233NLYQMTSQLE* | 23 | 68 | 3 | 28 | 0 | 0 | |
| 238-246WNQMNLGAT | 19 | 0.3 | 0 | 21 | 1 | 19 | |
| 242-250NLGATLKGV | 24 | 160 | 1 | 14 | 2 | 19 | |
| 390-398RKFSRSDHL | 11 | 0.054 | 1 | 27 | ND | ND | |
| 398-406LKTHTTRTHT | 5 | 0.18 | 1 | 22 | ND | ND | |
| 436-445NMHQRNHTKL* | 20 | 15 | 4 | 32 | 0 | 0 | |
| A0203 | 243-252LGATLKGVAA | 19 | NA | 0 | 21 | ND | ND |
| A2402 | 239-248NQMNLGATL | 10 | 7.2 | 0 | 2 | 1 | 17 |
| 436-445NMHQRNHTKL* | 13 | 0.6 | 13 | 27 | 0 | 0 | |
| A6901 | 246-253TLKGVAAGS | NA | NA | 0 | 57 | ND | ND |
| B0702 | (−125)-(−117)RQRPHPGAL | 15 | 40 | 1 | 53 | 1 | 67 |
| (−119)-(−111)GALRNPTAC | 2 | 0.3 | 5 | 22 | 1 | 60 | |
| A3101 | (−107)-(−99)HFPPSLPPT | NA | 0.01 | 0 | 27 | ND | ND |
| B3501 | (−8)-(−1)ASGSEPQQM | NA | 15 | 3 | 51 | 5 | 39 |
| 135-143PSCLESQPA | NA | 0.075 | 0 | 21 | ND | ND | |
| 174-182HSFKHEDPM | NA | 10 | 3 | 63 | 5 | 45 | |
| 269-278GYESDNHTT | NA | 0.004 | 0 | 23 | ND | ND | |
| 323-332FMCAYPGCNK | NA | 0.01 | 0 | 61 | 5 | 45 | |
| B3503 | 122-130SGQARMFPN | NA | NA | 3 | 41 | ND | ND |
| 218-226RTPYSSDNL | NA | NA | 3 | 31 | 4 | 48 | |
| B3508 | 238-246WNQMNLGAT | NA | NA | 2 | 21 | 4 | 19 |
| B3802 | 206-214TDSCTGSQA | NA | NA | 1 | 53 | ND | ND |
| B3801 | 166-174HHAAQFPNH | 11 | 0.3 | 1 | 17 | ND | ND |
| B3901 | 30-38GAAQWAPVL | 12 | 3 | 0 | 19 | ND | ND |
| B4001 | (−99)-(−91)THSPTHPPR | 3 | 0.02 | 0 | 31 | 3 | 65 |
| 46-54SAYGSLGGP | 1 | 0.002 | 8 | 24 | 3 | 68 | |
| 436-445NMHQRNHTKL | 1 | 0.002 | 1 | 26 | 3 | 72 | |
| B4402 | 202-210CHTPTDSCT | 3 | NA | 7 | 19 | ND | ND |
| 206-214TDSCTGSQA | 2 | NA | 0 | 88 | 1 | 56 | |
| 106-114ACRYGPFGP | 4 | NA | 7 | 23 | ND | ND | |
| B4701 | (−47)-(−37)PGCLQQPEQ | 1 | NA | 1 | 25 | ND | ND |
| B5701 | 6-15RDLNALLPAV | NA | NA | 1 | 22 | ND | ND |
| C0401 | 122-130SGQARMFPN | NA | NA | 3 | 41 | ND | ND |
| C1701 | 238-246WNQMNLGAT | NA | NA | 0 | 7 | 1 | 16 |
| DRB10101 | (−47)-(−37)PGCLQQPEQQG | 8 | NA | 1 | 25 | ND | ND |
| DRB10402 | 38-48LDFAPPGASAY | NA | NA | 1 | 71 | 0 | 40 |
| DRB10402 | 235-249CMTWNQMNLGATLKG | NA | NA | 2 | 15 | 1 | 17 |
| DRB10401 | 320-334KRPFMCAYPGC | 22 | NA | 3 | 0 | 5 | 5 |
| DRB11104 | 238-248WNQMNLGATLK | NA | NA | 2 | 1 | 0 | 0 |
| Presenting HLA allele . | Sequence identified . | Prediction algorithm . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs: . | ||||
|---|---|---|---|---|---|---|---|
| Binding index . | Disassociation time . | WT-1− allo-APCs with restricting HLA allele . | WT-1+ allo-APCs with restricting HLA allele loaded with WT-1 peptide . | WT-1− leukemia cells . | WT-1+ leukemia cells . | ||
| A0101 | 146-154NQGYSTVTF | 3 | 0.001 | 4 | 15 | ND | ND |
| 209-217CTGSQALLL | 12 | 0.125 | 0 | 26 | 3 | 33 | |
| 238-246WNQMNLGAT | 2 | 0 | 3 | 19 | ND | ND | |
| 242-250NLGATLKGV | 3 | 0.01 | 1 | 17 | ND | ND | |
| 269-278GYESDNHTT | 15 | 1.5 | 0 | 26 | 0 | 33 | |
| 323-332FMCAYPGCNK* | 0 | 0.1 | 2 | 0 | 5 | 0 | |
| A0201 | (−110)-(−102)PLPHFPPSL | 21 | 2 | 1 | 24 | 2 | 22 |
| (−99)-(−91)THSPTHPPR | 3 | 0 | 1 | 21 | 0 | 38 | |
| (−75)-(−67)AILDFLLLQ | 19 | 0.272 | 3 | 17 | 3 | 19 | |
| (−47)-(−39)PGCLQQPEQ | 0 | 0 | 7 | 27 | 5 | 19 | |
| (−27)-(−19)KLGAAEASA | 19 | 17 | 2 | 22 | 10 | 37 | |
| 6-15RDLNALLPAV | 18 | 0.2 | 4 | 31 | 0 | 9 | |
| 22-31GGCALPVSGA | 13 | 0.003 | 3 | 25 | 3 | 47 | |
| 38-46LDFAPPGAS | 11 | 0 | 1 | 62 | 0 | 40 | |
| 46-54SAYGSLGGP* | 14 | 0 | 5 | 31 | 0 | 0 | |
| 58-66PAPPPPPPP* | 5 | 0 | 1 | 18 | 0 | 0 | |
| 126-134RMFPNAPYL† | 22 | 313 | 1 | 52 | 2 | 25 | |
| 225-233NLYQMTSQLE* | 23 | 68 | 3 | 28 | 0 | 0 | |
| 238-246WNQMNLGAT | 19 | 0.3 | 0 | 21 | 1 | 19 | |
| 242-250NLGATLKGV | 24 | 160 | 1 | 14 | 2 | 19 | |
| 390-398RKFSRSDHL | 11 | 0.054 | 1 | 27 | ND | ND | |
| 398-406LKTHTTRTHT | 5 | 0.18 | 1 | 22 | ND | ND | |
| 436-445NMHQRNHTKL* | 20 | 15 | 4 | 32 | 0 | 0 | |
| A0203 | 243-252LGATLKGVAA | 19 | NA | 0 | 21 | ND | ND |
| A2402 | 239-248NQMNLGATL | 10 | 7.2 | 0 | 2 | 1 | 17 |
| 436-445NMHQRNHTKL* | 13 | 0.6 | 13 | 27 | 0 | 0 | |
| A6901 | 246-253TLKGVAAGS | NA | NA | 0 | 57 | ND | ND |
| B0702 | (−125)-(−117)RQRPHPGAL | 15 | 40 | 1 | 53 | 1 | 67 |
| (−119)-(−111)GALRNPTAC | 2 | 0.3 | 5 | 22 | 1 | 60 | |
| A3101 | (−107)-(−99)HFPPSLPPT | NA | 0.01 | 0 | 27 | ND | ND |
| B3501 | (−8)-(−1)ASGSEPQQM | NA | 15 | 3 | 51 | 5 | 39 |
| 135-143PSCLESQPA | NA | 0.075 | 0 | 21 | ND | ND | |
| 174-182HSFKHEDPM | NA | 10 | 3 | 63 | 5 | 45 | |
| 269-278GYESDNHTT | NA | 0.004 | 0 | 23 | ND | ND | |
| 323-332FMCAYPGCNK | NA | 0.01 | 0 | 61 | 5 | 45 | |
| B3503 | 122-130SGQARMFPN | NA | NA | 3 | 41 | ND | ND |
| 218-226RTPYSSDNL | NA | NA | 3 | 31 | 4 | 48 | |
| B3508 | 238-246WNQMNLGAT | NA | NA | 2 | 21 | 4 | 19 |
| B3802 | 206-214TDSCTGSQA | NA | NA | 1 | 53 | ND | ND |
| B3801 | 166-174HHAAQFPNH | 11 | 0.3 | 1 | 17 | ND | ND |
| B3901 | 30-38GAAQWAPVL | 12 | 3 | 0 | 19 | ND | ND |
| B4001 | (−99)-(−91)THSPTHPPR | 3 | 0.02 | 0 | 31 | 3 | 65 |
| 46-54SAYGSLGGP | 1 | 0.002 | 8 | 24 | 3 | 68 | |
| 436-445NMHQRNHTKL | 1 | 0.002 | 1 | 26 | 3 | 72 | |
| B4402 | 202-210CHTPTDSCT | 3 | NA | 7 | 19 | ND | ND |
| 206-214TDSCTGSQA | 2 | NA | 0 | 88 | 1 | 56 | |
| 106-114ACRYGPFGP | 4 | NA | 7 | 23 | ND | ND | |
| B4701 | (−47)-(−37)PGCLQQPEQ | 1 | NA | 1 | 25 | ND | ND |
| B5701 | 6-15RDLNALLPAV | NA | NA | 1 | 22 | ND | ND |
| C0401 | 122-130SGQARMFPN | NA | NA | 3 | 41 | ND | ND |
| C1701 | 238-246WNQMNLGAT | NA | NA | 0 | 7 | 1 | 16 |
| DRB10101 | (−47)-(−37)PGCLQQPEQQG | 8 | NA | 1 | 25 | ND | ND |
| DRB10402 | 38-48LDFAPPGASAY | NA | NA | 1 | 71 | 0 | 40 |
| DRB10402 | 235-249CMTWNQMNLGATLKG | NA | NA | 2 | 15 | 1 | 17 |
| DRB10401 | 320-334KRPFMCAYPGC | 22 | NA | 3 | 0 | 5 | 5 |
| DRB11104 | 238-248WNQMNLGATLK | NA | NA | 2 | 1 | 0 | 0 |
Epitopes are listed according to their presenting HLA alleles.
NA indicates not available; and ND, not determined.
T cells cytotoxic against the autologous WT-1 peptide–loaded APCs but not the leukemic cells.
Previously reported epitopes.
As can be seen in Table 1, of 51 cultures generating IFNγ+ CD8+ T cells after secondary stimulation with an identified peptide-loaded autologous APCs, 50 also exhibited significant specific cytotoxic activity against autologous PHA blasts loaded with the targeted peptide. Of these, 48 also lysed allogeneic peptide–loaded PHA blasts or DCs sharing the restricting HLA allele of the responding T cells. CD4+ IFNγ+ T cells responding to 3 of 5 identified 11-mer peptides presented by class II HLA alleles also lysed peptide-loaded autologous and HLA-sharing allogeneic class II+ targets.
Of the T-cell cultures exhibiting epitope-specific cytotoxic activity against peptide-loaded targets, 36 could be tested for cytotoxic activity against WT-1+ leukemic cells coexpressing the T cells' restricting HLA allele. Of these 36, 27 exhibited HLA-restricted cytotoxic activity against the WT-1+ leukemic cells (Table 2). T cells specific for 5 peptides, 6-15RDL, 46-54SAY, 58-66PAP, 225-233NLY, and 436-445NMH, presented by HLA-A0201, could not lyse HLA-A0201+ WT-1+ leukemic cells. However, HLA-B4001–restricted T cells specific for the 46-54SAY peptide could lyse WT-1+ leukemic cells coexpressing this HLA allele. Similarly, NMH peptide–specific HLA-restricted T-cell lines that lysed targets loaded with the NMH peptide coexpressing HLA-A0201, HLA-B4001, or HLA-A2402 were only able to lyse WT-1+ leukemic cells expressing the HLA-B4001 allele.
To ascertain that the cytotoxic activity of the WT-1 peptide–specific T cells observed against allogeneic WT-1+ leukemic cells sharing the T-cells' restricting allele did not reflect the presence of alloresponsive T cells in the T-cell lines, we tested the cytotoxic activity of 13 of these HLA-restricted WT-1 peptide–specific T-cell lines against WT-1+ leukemic cells and WT-1− PHA blasts cultured from the same leukemic patient. As shown in Table 3, the WT-1–specific T cells lysed the WT-1+ leukemic cells but not PHA blasts from the same patient. We did not have PHA blasts from every patient who provided leukemia blasts for this study. Nevertheless, these results provide evidence that the cytotoxicity of the WT-1–specific T cells is not ascribable to contaminating alloreactivity. A second, more inclusive but less direct line of evidence is provided by a paired comparison of the responses of T cells derived from 35 of the donors who had been contemporaneously sensitized in vitro against either WT-1 peptide pool loaded or unmodified autologous EBV-BLCLs against these primary leukemias. As shown in Table 4, T cells sensitized with the WT-1 peptide pool–loaded EBV-BLCLs lysed WT-1+ leukemic cells sharing the T cells' restricting HLA allele in 25 of 35 cases. In contrast, T cells sensitized with autologous EBV-BLCLs alone consistently failed to lyse the same WT-1+ leukemia targets.
Cytotoxic activity of T cells specific for WT-1–derived immunogenic epitopes identified by IFNγ production assay after sensitization with the pool of overlapping pentadecapeptides spanning the sequence of the WT-1 protein when tested against WT-1+ primary leukemic cells and PHA blasts of the same origin (P < .001)
| 15-mer no. containing the dominant epitope . | Sequence identified . | Presenting HLA allele . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs: . | |
|---|---|---|---|---|
| WT-1+ leukemia cells* . | PHA blasts† . | |||
| 1 | (−125)-(−117)RQRPHPGAL | B0702 | 67 | 2 |
| 2 | (−119)-(−111)GALRNPTAC | B0702 | 60 | 1 |
| 4 | (−110)-(−102)PLPHFPPSL | A0201 | 22 | 1 |
| 7 | (−99)-(−91)THSPTHPPR | B4001 | 65 | 5 |
| A0201 | 38 | 3 | ||
| 24-25 | (−27)-(−19)KLGAAEASA | A0201 | 37 | 8 |
| 29-30 | (−8)-(1)ASGSEPQQM | B3501 | 39 | 9 |
| 37 | 22-31GGCALPVSGA | A0201 | 47 | 6 |
| 43 | 46-54SAYGSLGGP | B4001 | 68 | 3 |
| 62-63 | 126-134RMFPNAPYL‡ | A0201 | 25 | 3 |
| 86 | 218-226RTPYSSDNL | B3503 | 48 | 1 |
| C0401 | 48 | 1 | ||
| 141 | 436-445NMHQRNHTKL | B4001 | 72 | 1 |
| 15-mer no. containing the dominant epitope . | Sequence identified . | Presenting HLA allele . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs: . | |
|---|---|---|---|---|
| WT-1+ leukemia cells* . | PHA blasts† . | |||
| 1 | (−125)-(−117)RQRPHPGAL | B0702 | 67 | 2 |
| 2 | (−119)-(−111)GALRNPTAC | B0702 | 60 | 1 |
| 4 | (−110)-(−102)PLPHFPPSL | A0201 | 22 | 1 |
| 7 | (−99)-(−91)THSPTHPPR | B4001 | 65 | 5 |
| A0201 | 38 | 3 | ||
| 24-25 | (−27)-(−19)KLGAAEASA | A0201 | 37 | 8 |
| 29-30 | (−8)-(1)ASGSEPQQM | B3501 | 39 | 9 |
| 37 | 22-31GGCALPVSGA | A0201 | 47 | 6 |
| 43 | 46-54SAYGSLGGP | B4001 | 68 | 3 |
| 62-63 | 126-134RMFPNAPYL‡ | A0201 | 25 | 3 |
| 86 | 218-226RTPYSSDNL | B3503 | 48 | 1 |
| C0401 | 48 | 1 | ||
| 141 | 436-445NMHQRNHTKL | B4001 | 72 | 1 |
Differences in responses to WT-1+ leukemia cells and PHA blasts, analysed by paired ttest, are significant (P < .001).
Leukemia targets were derived from either immortalized leukemia cell lines or primary leukemia cells obtained from patients with WT-1+ leukemia.
PHA blasts were generated from PBMCs derived from the same patients as the WT-1+ primary leukemia cells.
Epitopes previously predicted by the computer algorithm or described in the literature.
Leukemocidal activity of defined epitope-specific and HLA-restricted T cells from normal donors sensitized with either autologous EBV-BLCLs or EBV-BLCLs loaded with pooled WT-1 peptides against primary WT-1+ leukemia cells sharing the T cells' restricting HLA allele (P < .001)
| 15-mer no. Containing the dominant epitope . | Sequence identified . | Presenting HLA allele . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs WT-1+ leukemia cells expressing restricting HLA allele . | |
|---|---|---|---|---|
| WT-1 CTLs . | EBV CTLs . | |||
| 1 | (−125)-(−117)RQRPHPGAL | B0702 | 67 | 1 |
| 2 | (−119)-(−111)GALRNPTAC | B0702 | 60 | 2 |
| 4 | (−110)-(−102)PLPHFPPSL | A0201 | 22 | 3 |
| 7 | (−99)-(−91)THSPTHPPR | B4001 | 65 | 0 |
| A0201 | 38 | 3 | ||
| 13 | (−75)-(67)AILDFLLLQ | A0201 | 19 | 5 |
| 20 | (−47)-(39)PGCLQQPEQ | A0201 | 19 | 10 |
| 24-25 | (−27)-(−19)KLGAAEASA | A0201 | 37 | 5 |
| 29-30 | (−8)-(1)ASGSEPQQM | B3501 | 39 | 0 |
| 33 | 6-15RDLNALLPAV | A0201 | 9 | 0 |
| 37 | 22-31GGCALPVSGA | A0201 | 47 | 3 |
| 41 | 38-46LDFAPPGAS | A0201 | 40 | 0 |
| 38-48LDFAPPGASAY | DRB10402 | 40 | 0 | |
| 43 | 46-54SAYGSLGGP | A0201 | 0 | 0 |
| B4001 | 68 | 3 | ||
| 46 | 58-66PAPPPPPPP | A0201 | 0 | 0 |
| 62-63 | 126-134RMFPNAPYL* | A0201 | 25 | 2 |
| 74-75 | 174-182HSFKHEDPM | B3501 | 45 | 5 |
| 83-84 | 209-217CTGSQALLL | A0101 | 33 | 3 |
| 83 | 206-214TDSCTGSQA | B4402 | 56 | 1 |
| 86 | 218-226RTPYSSDNL | B3503 | 48 | 4 |
| C0401 | 48 | 4 | ||
| 87 | 225-233NLYQMTSQLE | A0201 | 0 | 0 |
| 91 | 238-246WNQMNLGAT | A0201 | 19 | 1 |
| C1701 | 16 | 1 | ||
| B3508 | 19 | 4 | ||
| 91-92 | 239-248NQMNLGATL | A2402 | 17 | 1 |
| 91 | 238-248WNQMNLGATLK | DRB11104 | 0 | 0 |
| 235-249CMTWNQMNLGATLKG | DRB10402 | 17 | 1 | |
| 92 | 242-250NLGATLKGV | A0201 | 19 | 2 |
| 99-100 | 269-278GYESDNHTT | A0101 | 33 | 0 |
| 112-113 | 323-332FMCAYPGCNK | B3501 | 45 | 5 |
| 320-334KRPFMCAYPGC | DRB10401 | 5 | 5 | |
| 141 | 436-445NMHQRNHTKL* | A0201 | 0 | 0 |
| B4001 | 72 | 3 | ||
| A2402 | 0 | 0 | ||
| 15-mer no. Containing the dominant epitope . | Sequence identified . | Presenting HLA allele . | Cytotoxic CTL response, % (at a 50:1 effector: stimulator ratio) vs WT-1+ leukemia cells expressing restricting HLA allele . | |
|---|---|---|---|---|
| WT-1 CTLs . | EBV CTLs . | |||
| 1 | (−125)-(−117)RQRPHPGAL | B0702 | 67 | 1 |
| 2 | (−119)-(−111)GALRNPTAC | B0702 | 60 | 2 |
| 4 | (−110)-(−102)PLPHFPPSL | A0201 | 22 | 3 |
| 7 | (−99)-(−91)THSPTHPPR | B4001 | 65 | 0 |
| A0201 | 38 | 3 | ||
| 13 | (−75)-(67)AILDFLLLQ | A0201 | 19 | 5 |
| 20 | (−47)-(39)PGCLQQPEQ | A0201 | 19 | 10 |
| 24-25 | (−27)-(−19)KLGAAEASA | A0201 | 37 | 5 |
| 29-30 | (−8)-(1)ASGSEPQQM | B3501 | 39 | 0 |
| 33 | 6-15RDLNALLPAV | A0201 | 9 | 0 |
| 37 | 22-31GGCALPVSGA | A0201 | 47 | 3 |
| 41 | 38-46LDFAPPGAS | A0201 | 40 | 0 |
| 38-48LDFAPPGASAY | DRB10402 | 40 | 0 | |
| 43 | 46-54SAYGSLGGP | A0201 | 0 | 0 |
| B4001 | 68 | 3 | ||
| 46 | 58-66PAPPPPPPP | A0201 | 0 | 0 |
| 62-63 | 126-134RMFPNAPYL* | A0201 | 25 | 2 |
| 74-75 | 174-182HSFKHEDPM | B3501 | 45 | 5 |
| 83-84 | 209-217CTGSQALLL | A0101 | 33 | 3 |
| 83 | 206-214TDSCTGSQA | B4402 | 56 | 1 |
| 86 | 218-226RTPYSSDNL | B3503 | 48 | 4 |
| C0401 | 48 | 4 | ||
| 87 | 225-233NLYQMTSQLE | A0201 | 0 | 0 |
| 91 | 238-246WNQMNLGAT | A0201 | 19 | 1 |
| C1701 | 16 | 1 | ||
| B3508 | 19 | 4 | ||
| 91-92 | 239-248NQMNLGATL | A2402 | 17 | 1 |
| 91 | 238-248WNQMNLGATLK | DRB11104 | 0 | 0 |
| 235-249CMTWNQMNLGATLKG | DRB10402 | 17 | 1 | |
| 92 | 242-250NLGATLKGV | A0201 | 19 | 2 |
| 99-100 | 269-278GYESDNHTT | A0101 | 33 | 0 |
| 112-113 | 323-332FMCAYPGCNK | B3501 | 45 | 5 |
| 320-334KRPFMCAYPGC | DRB10401 | 5 | 5 | |
| 141 | 436-445NMHQRNHTKL* | A0201 | 0 | 0 |
| B4001 | 72 | 3 | ||
| A2402 | 0 | 0 | ||
Differences between responses of T cells sensitized with WT-1 peptide pool loaded EBVBLCL and those sensitized with EBVBLCL alone, analyzed by paired t test, are significant (P < .001).
Epitopes previously predicted by the computer algorithm or described in the literature.
Immunogenicity of the newly identified WT-1 epitopes
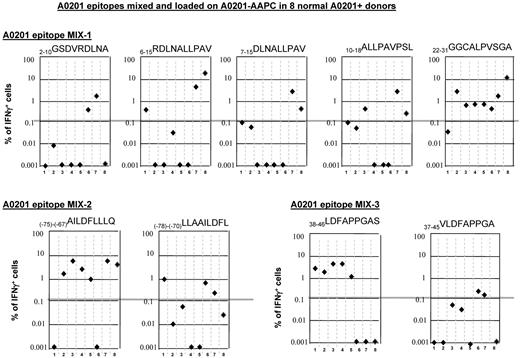
To ascertain that that the peptides identified by mapping responses in single donors were also immunogenic in a high proportion of individuals bearing the same presenting HLA allele, we investigated whether these epitopes could elicit appropriately restricted T-cell responses in groups of 6-12 individuals expressing that HLA allele. For this purpose, the T cells from each donor were sensitized with the identified epitope loaded on a panel of AAPCs,42 each expressing a single HLA allele, specifically A0201, A0301, A2402, or B0702. As shown in Table 5, of 9 peptides identified that are presented by HLA-A0201, all were able to stimulate WT-1–specific IFNγ+ T-cell responses in a proportion of HLA-A0201+ individuals. The previously reported 126-134RMFPNAPYL peptide presented by HLA-A0201 allele elicited responses in 5 of 12 (42%) HLA-A0201+ healthy donors tested. In comparison, 5 of the other 8 peptides tested elicited WT-1 peptide–specific responses in 50%-75% of the same HLA-A0201+ donors. Two WT-1 epitopes presented by the HLA-B0702 allele also elicited WT-1–specific T-cell responses in 50% and 63% of the tested individuals, respectively (Table 5). All of the peptides tested elicited specific responses in at least 2 additional donors bearing their presenting HLA allele.
Proportion of normal donors generating WT-1 peptide–specific T-cell responses after stimulation with AAPCs expressing preselected restricting HLA alleles loaded with specific WT-1–derived immunogenic epitopes identified previously by mapping IFNγ+ T-cell responses to a pool of overlapping pentadecapeptides spanning the whole sequence of the WT-1 protein
| Presenting HLA allele . | Sequence identified . | Identified in no. of donors after total pool stimulation on CAMs . | Proportion of responses in healthy donors to the peptide stimulation (%) . | Predicted . | Predicted sequence . | Predicted . | Proportion of responses in healthy donors to the peptide stimulation . | ||
|---|---|---|---|---|---|---|---|---|---|
| Binding index . | Dissociation time . | Binding index . | Dissociation time . | ||||||
| A0201 | (−99)-(−91)THSPTHPPR | 1 | 6/12 (50%) | 3 | 0 | (−99)-(−91)THSPTHPPR | 3 | 0 | 6/12 (50%) |
| (−75)-(−67)AILDFLLLQ | 1 | 8/12 (67%) | 19 | 0.272 | (−78)-(−70)LLAAILDFL | 28 | 225 | 8/12 (67%) | |
| (−47)-(−39)PGCLQQPEQ | 2 | 2/12 (16%) | 0 | 0 | (−45)-(−36)CLQQPEQQGV | 21 | 70 | 2/12 (16%) | |
| (−27)-(−19)KLGAAEASA | 1 | 8/12 (67%) | 19 | 17 | (−27)-(−19)KLGAAEASA | 19 | 17 | 8/12 (67%) | |
| 6-15RDLNALLPAV | 1 | 3/12 (25%) | 18 | 0.2 | 7-15DLNALLPAV | 27 | 12 | 3/12 (25%) | |
| 22-31GGCALPVSGA | 3 | 9/12 (75%) | 13 | 0.003 | 10-18ALLPAVPSL | 33 | 181 | 3/12 (25%) | |
| 38-46LDFAPPGAS | 2 | 8/12 (67%) | 11 | 0 | 37-45VLDFAPPGA | 16 | 4 | 0/12 (0%) | |
| 126-134RMFPNAPYL | 1 | 5/12 (42%) | 22 | 313 | 126-134RMFPNAPYL | 22 | 313 | 5/12 (42%) | |
| 238-246WNQMNLGAT | 2 | 3/12 (25%) | 19 | 0.3 | 235-243CMTWNQMNL | 17 | 1.5 | 0/8 | |
| Total pool | 13/27 (48%) | 8/12 (67%) | |||||||
| A2402 | 239-248NQMNLGATL | 1 | 4/6 (60%) | 10 | 7.2 | 235-243CMTWNQMNL | 10 | 4 | 1/6 (17%) |
| Total pool | 2/6 (33%) | 6/6 (100%) | |||||||
| B0702 | (−125)-(−117)RQRPHPGAL | 1 | 4/8 (50%) | 15 | 40 | (−125)-(−117)RQRPHPGAL | 15 | 40 | 4/8 (50%) |
| (−119)-(−111)GALRNPTAC | 1 | 5/8 (63%) | 2 | 0.3 | (−118)-(−109)ALRNPTACPL | 15 | 120 | 5/8 (63%) | |
| 323-332FMCAYPGCNK | 1 | 3/8 (38%) | 1 | 0.015 | 327-335YPGCNKRYF | 17 | 0.4 | 4/8 (50%) | |
| Total pool | 2/8 (25%) | 3/8 (38%) | |||||||
| Presenting HLA allele . | Sequence identified . | Identified in no. of donors after total pool stimulation on CAMs . | Proportion of responses in healthy donors to the peptide stimulation (%) . | Predicted . | Predicted sequence . | Predicted . | Proportion of responses in healthy donors to the peptide stimulation . | ||
|---|---|---|---|---|---|---|---|---|---|
| Binding index . | Dissociation time . | Binding index . | Dissociation time . | ||||||
| A0201 | (−99)-(−91)THSPTHPPR | 1 | 6/12 (50%) | 3 | 0 | (−99)-(−91)THSPTHPPR | 3 | 0 | 6/12 (50%) |
| (−75)-(−67)AILDFLLLQ | 1 | 8/12 (67%) | 19 | 0.272 | (−78)-(−70)LLAAILDFL | 28 | 225 | 8/12 (67%) | |
| (−47)-(−39)PGCLQQPEQ | 2 | 2/12 (16%) | 0 | 0 | (−45)-(−36)CLQQPEQQGV | 21 | 70 | 2/12 (16%) | |
| (−27)-(−19)KLGAAEASA | 1 | 8/12 (67%) | 19 | 17 | (−27)-(−19)KLGAAEASA | 19 | 17 | 8/12 (67%) | |
| 6-15RDLNALLPAV | 1 | 3/12 (25%) | 18 | 0.2 | 7-15DLNALLPAV | 27 | 12 | 3/12 (25%) | |
| 22-31GGCALPVSGA | 3 | 9/12 (75%) | 13 | 0.003 | 10-18ALLPAVPSL | 33 | 181 | 3/12 (25%) | |
| 38-46LDFAPPGAS | 2 | 8/12 (67%) | 11 | 0 | 37-45VLDFAPPGA | 16 | 4 | 0/12 (0%) | |
| 126-134RMFPNAPYL | 1 | 5/12 (42%) | 22 | 313 | 126-134RMFPNAPYL | 22 | 313 | 5/12 (42%) | |
| 238-246WNQMNLGAT | 2 | 3/12 (25%) | 19 | 0.3 | 235-243CMTWNQMNL | 17 | 1.5 | 0/8 | |
| Total pool | 13/27 (48%) | 8/12 (67%) | |||||||
| A2402 | 239-248NQMNLGATL | 1 | 4/6 (60%) | 10 | 7.2 | 235-243CMTWNQMNL | 10 | 4 | 1/6 (17%) |
| Total pool | 2/6 (33%) | 6/6 (100%) | |||||||
| B0702 | (−125)-(−117)RQRPHPGAL | 1 | 4/8 (50%) | 15 | 40 | (−125)-(−117)RQRPHPGAL | 15 | 40 | 4/8 (50%) |
| (−119)-(−111)GALRNPTAC | 1 | 5/8 (63%) | 2 | 0.3 | (−118)-(−109)ALRNPTACPL | 15 | 120 | 5/8 (63%) | |
| 323-332FMCAYPGCNK | 1 | 3/8 (38%) | 1 | 0.015 | 327-335YPGCNKRYF | 17 | 0.4 | 4/8 (50%) | |
| Total pool | 2/8 (25%) | 3/8 (38%) | |||||||
Comparison of responses to peptides identified by mapping responses to pooled WT-1 15-mers with responses to previously reported WT-1 peptides predicted by binding algorithms to be immunogenic
We also compared primary responses by healthy donor T cells to individual WT-1 peptides identified by our mapping strategy to responses against other WT-1 peptides containing flanking sequences predicted to have a higher binding index for the presenting HLA allele using binding algorithms described previously.44,45 As shown in Table 5, the predicted binding indices for 8 of 12 mapped epitopes were only somewhat lower than those for the most studied WT-1 peptide, RMF, presented by HLA-A0201. However, their dissociation times were markedly lower. Nevertheless, T-cell responses to each of these peptides were elicited in a high proportion of healthy donors.
In 5 cases, the mapped peptide specificity [ie, (−99)-(−91)THS, (−22)-(−19)KLG, 22-31GGC, 126-134RMF, and (−125)-(−117)RQR] was identical to the peptide with the highest affinity for the presenting HLA allele predicted by the binding algorithm within the stimulating 15-mer. In those cases in which the mapped sequences and the sequences predicted to have the highest binding index differed, the proportion of donors responding to individual mapped peptides were equal to or greater than those generated in response to the neighboring epitopes predicted to have higher affinity. For example, IFNγ+ T-cell responses were generated to the 38-46LDF peptide in 8 of 12 (67%) of HLA-A0201 donors tested, whereas none responded to the predicted and previously reported46 epitope 37-45VLDFAPPGA. Similarly, among HLA-A2402+ donors, 4 of 6 donors (60%) responded to the 239-248NQMNLGATL peptide, whereas only 1 of 6 responded to the 235-243CMTWNQMNL peptide previously reported to be presented by this allele.29
To compare directly peptides presented by HLA-A0201 that we identified by matrix mapping with flanking peptides with higher predicted binding indices, the peptides, mixed at equal concentrations, were loaded on HLA-A0201+ AAPCs and used to sensitize T cells from 8 of the HLA-A0201+ healthy donors. After 35 days of sensitization, the T cells were then washed and secondarily restimulated for 24 hours with aliquots of irradiated autologous PBMCs loaded with each individual peptide. Responding IFNγ+ T cells were then quantitated by FACS. The results, presented in Figure 4, demonstrate that although the 22-31GGC peptide had the lowest binding index and the shortest predicted dissociation time, it induced strong IFNγ+ T-cell responses in 7 of 8 donors. Furthermore, although 3 of 8 donors responded to the 6-15RDL, 10-18ALL and 7-15DLN peptides, 6-15RDL peptides identified by response mapping elicited higher numbers of IFNγ+ T cells. In comparisons of the (−75)-(−67)AILDFLLLQ with the flanking (−78)-(−70)LLAAILDFL sequence, the AIL peptide elicited superior responses and in a higher proportion of donors (6 of 8 vs 3 of 8 donors, respectively). Similarly, in comparisons of the mapped 38-46LDFAPPGAS peptide with the previously reported 37-45VLDFAPPGA peptide,46 the LDF peptide induced strong responses in 5 of the 8 donors, whereas the VLD peptide induced low responses in only 2 of these donors.
Discussion
Among those antigens uniquely or differentially expressed by malignant cells, WT-1 is considered to be one of the most promising.47 However, the number of immunogenic WT-1 peptide antigens previously identified and reported is very limited and largely confined to a set of peptides presented by the HLA alleles A0201, A2402, and DRB10401. Using a pool of overlapping 15-mer peptides spanning the amino acid sequence of WT-1 loaded on autologous APCs for sensitization, we were able to generate WT-1 peptide–specific IFNγ+CD4+ and CD8 T-cell responses from the blood of 41 of 56 (78%) healthy donors and to thereafter identify the epitopes eliciting these responses and their presenting HLA alleles. Of the 42 WT-1 peptide antigens defined, all but one have not been described previously. The new immunogenic peptides identified include 36 peptides presented by class I HLA alleles and 5 presented by class II HLA alleles. Of the peptides presented by class I HLA alleles, 10 nonamer epitopes were identified that could be presented by 2-4 different HLA alleles. We also identified, within 4 pentadecapeptides, overlapping 11-mer and nonamer sequences that coinduced distinguishable CD4+ IFNγ+ and CD8+ IFNγ+ T cells. Whether and to what degree epitopes that can be presented by more than one allele can elicit enhanced WT-1 specific responses in individuals inheriting both presenting HLA alleles, or both the class I and class II presenting HLA alleles in those instances in which overlapping sequences are contained in the same 15-mer, is as yet unclear. However, inclusion of such peptides in WT-1 vaccines could significantly broaden their applicability, particularly among patients not inheriting HLA-A0201 or HLA-A2402.
Those peptides presented by class I HLA alleles elicited IFNγ+ CD8+ T cells that were able to lyse peptide-loaded autologous APCs and allogeneic APCs sharing the T cells' restricting HLA allele in 50 of 51 (99%) and 48 of 51 (94%) cultures tested, respectively (Tables 1 and 2). More importantly, of 36 HLA-restricted WT-1 peptide–specific T-cell lines that could be tested, T-cell lines specific for 29 epitopes, including 2 of 4 epitopes presented by class II and 27 of 32 presented by class I alleles, were also able to lyse WT-1+ leukemic blasts sharing the T cells' restricting HLA allele. The failure of the HLA-restricted WT-1 epitope–specific T cells to lyse allogeneic PHA blasts from the same leukemic patients (Table 3), coupled with the differential leukemocidal activity of T cells sensitized with WT-1 peptide–loaded autologous EBV-BLCLs compared with aliquots of the same T cells sensitized with autologous EBV-BLCLs alone (Table 4), indicates that the leukemocidal activity is WT-1 peptide–specific and not a result of contaminating alloreactive T cells. Therefore, our data suggest that 29 of 36 immunogenic peptides of WT-1 identified (80%) can be processed and presented by WT-1+ leukemic cells at concentrations adequate for WT-1 epitope–specific T-cell recognition and cytolysis.
An unexpected result of our epitope-mapping studies was the lack of responses specific for previously reported WT-1 peptide antigens other than the 126-134RMF peptide. For example, no T-cell responses were recorded against the 187-195SLG,30 37-45VLD,46 or 10-19ALL35 peptides when HLA-A0201+ T cells were sensitized with the complete pool of WT-1, nor were HLA-A2402–restricted responses generated against the 215-243CMT29 peptide, even though each of these peptides is contained in single 15-mers that are a part of the pool. Fine mapping of the specific nonamers eliciting responses identified alternative peptides in these 15-mers, usually containing part of the sequences predicted to elicit responses on the basis of HLA-binding algorithms, as the immunogenic epitopes. A striking example is the 37-45VLD peptide previously reported to be immunogenic based on binding algorithms when presented by HLA-A0201.46 This peptide did not elicit a response in any of the 12 HLA-A0201+ donors when their T cells were sensitized with the whole pool. In contrast, the overlapping 38-46LDF peptide induced specific HLA-A0201–restricted IFNγ+ T cells that were also leukemocidal in T cells cultured from 8 of 12 HLA-A0201+ donors (Table 5). In certain cases, responses to a specific nonamer sequence within a 15-mer might be altered by differences in the susceptibility of amino acid sequences flanking the nonamer to editing by exoproteases.48 However, in direct comparisons, the 38-46LDF nonamer peptide also elicited stronger and more consistent T-cell responses (Figure 4). This also held true in other direct comparisons of the immunogenicity of peptides when presented by AAPCs selectively expressing HLA-A0201. Therefore, as has also been shown for viral epitopes,49 the capacity of a given peptide to elicit a T-cell response is not consistently correlated with its predicted potential to bind its presenting HLA allele.
IFNγ+ T-cell responses to equimolar mixtures of 9-mer peptides identified by epitope mapping of in vitro responses and peptides within the same 15-mer or adjacent overlapping 15-mer peptides predicted to have higher binding affinity and immunogenicity. (A) Responses to a mixture of nonamers spanning amino acids +2 to +31 including the 6-15RDL and 22-31GGC peptides to which HLA A0201+ donors responded in epitope-mapping studies. (B) Responses to the in vitro–mapped (−75)-(−67)AILDFLLLQ epitope and a flanking peptide (−78)-(−70)LLAAILDFL with higher predicted binding affinity. (C) Responses to the in vitro–mapped 38-46LDFAPPGAS epitope and the overlapping 37-45VLDFAPPGA predicted to have higher binding affinity.
IFNγ+ T-cell responses to equimolar mixtures of 9-mer peptides identified by epitope mapping of in vitro responses and peptides within the same 15-mer or adjacent overlapping 15-mer peptides predicted to have higher binding affinity and immunogenicity. (A) Responses to a mixture of nonamers spanning amino acids +2 to +31 including the 6-15RDL and 22-31GGC peptides to which HLA A0201+ donors responded in epitope-mapping studies. (B) Responses to the in vitro–mapped (−75)-(−67)AILDFLLLQ epitope and a flanking peptide (−78)-(−70)LLAAILDFL with higher predicted binding affinity. (C) Responses to the in vitro–mapped 38-46LDFAPPGAS epitope and the overlapping 37-45VLDFAPPGA predicted to have higher binding affinity.
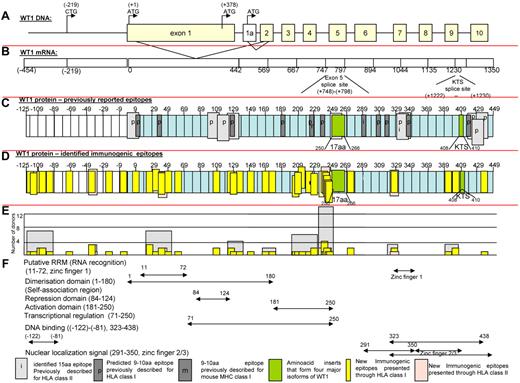
In Figure 5, we present maps of the WT-1 protein. Figure 5C defines the localization of all previously reported antigenic epitopes presented by HLA class I and II alleles; Figure 5D depicts the location of immunogenic peptides identified in this report. As can be seen, the 11 epitopes previously reported to be presented by class I and 10 presented by class II HLA alleles are principally clustered in sequences encoded by exons 1, 7, and 10, whereas the epitopes recognized by healthy T cells sensitized with the WT-1 peptide pool are principally clustered in sequences encoded by the first 5 exons. Therefore, 26 of the new epitopes are included in each of the 4 major isoforms of WT-1 resulting from splice variants that do or do not include the 17 amino acid sequence (amino acids 250-266) in exon 5 or the 3–amino acid sequence (400-410KTS) between zinc fingers 3 and 4. Whereas the epitopes are distributed broadly, clusters of epitopes were detected in the RNA recognition domain in exon 1 and the activation domain (amino acids 181-250; Figure 5F) proximal to the spliced 17–amino acid segment in exon 5. The latter area also contained those epitopes most frequently recognized by multiple donors (Figure 5E). Interestingly, 9 newly identified epitopes map to a 126–amino acid sequence at the N-terminus encoded by a segment of the WT-1 gene initially described by Gessler et al37 that is centromeric to exon 1 of the (exon 5+, KTS+) isoform of WT-1 and includes the long isoform of WT-1 initiated at a CUG codon upstream of the AUG initiator for exon 1.50 Strikingly, each of the epitopes identified in this sequence elicits IFNγ+ T cells that are cytolytic against leukemic blasts coexpressing WT-1 and the T cells' restricting HLA allele.
Schema of WT-1. (A) Schema of WT-1 DNA. (B) Schema of WT-1 mRNA (includes region encoding first 126 amino acids within the N-terminus reported by Gessler et al37 ). (C) Localization of the previously described immunogenic epitopes presented through HLA class I (dark gray) and HLA class II (light gray) either predicted (p) by the computer algorithm or identified (i) by the comparative analysis of the immunogenicity of a panel of neighboring peptides. The 126 amino acids at the N-terminus are numbered with negative values from the first AUG codon in exon 1. (D) Localization of the immunogenic epitopes identified in this study by the epitope-mapping approach presented by HLA class I (yellow) and HLA class II (pink) alleles and marked within the schema of the WT-1 protein 575 amino acids in length, including 126 amino acids labeled with negative numbers at the N-terminus upstream from the first AUG codon initiating exon 1. (E) Number of healthy donors responding to the identified epitopes (yellow) or cluster of epitopes (gray). (F) Distribution of the functional activities among the corresponding regions of the WT-1 protein.
Schema of WT-1. (A) Schema of WT-1 DNA. (B) Schema of WT-1 mRNA (includes region encoding first 126 amino acids within the N-terminus reported by Gessler et al37 ). (C) Localization of the previously described immunogenic epitopes presented through HLA class I (dark gray) and HLA class II (light gray) either predicted (p) by the computer algorithm or identified (i) by the comparative analysis of the immunogenicity of a panel of neighboring peptides. The 126 amino acids at the N-terminus are numbered with negative values from the first AUG codon in exon 1. (D) Localization of the immunogenic epitopes identified in this study by the epitope-mapping approach presented by HLA class I (yellow) and HLA class II (pink) alleles and marked within the schema of the WT-1 protein 575 amino acids in length, including 126 amino acids labeled with negative numbers at the N-terminus upstream from the first AUG codon initiating exon 1. (E) Number of healthy donors responding to the identified epitopes (yellow) or cluster of epitopes (gray). (F) Distribution of the functional activities among the corresponding regions of the WT-1 protein.
Of the several “self” proteins differentially expressed by specific tumors (eg, WT-1, NY-ESO-1, HER2/neu, MAGE, and others), only WT-1 and MART-1 have been shown to elicit responses in healthy donors.31,32,51–54 In contrast, T cells specific for each of these proteins have been recorded in a proportion of patients with tumors overexpressing them.55 In particular, T cells specific for the RMF and CMT peptides of WT-1 have been detected in patients with leukemias, myeloma, carcinoma of the breast and prostate, and other solid tumors.31,32,56–61 In an ongoing study, we have also documented responses to several of the WT-1 epitopes identified in the present study in 50%-60% of patients with ovarian cancer. Given the high number of potentially immunogenic epitopes in proteins such as NY-ESO-1 and HER2/neu that have elicited responses in tumor-bearing hosts,62 the number of immunogenic WT-1 peptides that we have identified is not sufficiently different to account for the differential presence of WT-1 responses in healthy donors. Furthermore, Pospori et al have shown that hematopoietic stem cells expressing a transduced TCR specific for a WT-1 peptide presented by HLA-A0201 are not deleted in the thymus of HLA-A0201–transgenic mice and generate functional memory T cells.63 However, whereas the basis for this lack of “self” tolerance is unclear, the studies of Rezvani et al31 and our own data (Figure 1A) indicate that the frequencies of WT-1–specific T cells in the blood of healthy donors is low. This may in part reflect the low levels and limited tissue distribution of WT-1 expression in healthy subjects.18–20 Recently, Rezvani et al also demonstrated declining T-cell responses to WT-1 in patients repeatedly vaccinated with WT-1 peptides,64 suggesting that these responses are highly regulated. Lehe et al have also recently shown that sensitization of T cells with a WT-1 peptide presented by DRB10402 in the presence of high concentrations of IL-2 preferentially stimulates the generation of CD25+ FOX P3+ GITR+ CD127− regulatory T cells capable of inhibiting CD8+ WT-1–specific T-cell responses.65
Under the culture conditions used in the present study, autologous dendritic cells and EBV-BLCLs loaded with the WT-1 peptide pool preferentially induced the generation of CD8+ and CD4+ IFNγ+ WT-1 peptide–specific T cells from 41 of 56 healthy donors (73%). Although each donor recognized only 1-3 epitopes of WT-1, the fact that T cells specific for 80% of these epitopes could recognize WT-1+ leukemic cells sharing the T cells' presenting HLA allele suggests that the turnover and processing of the aberrantly expressed WT-1 is high, permitting the simultaneous presentation of several different WT-1 epitopes by the restricting HLA allele expressed by these leukemic cells. Whether high expression of WT-1 or specific isoforms thereof permits differential presentation of specific epitopes, such as those in exon 5 or at the NH2 terminus of WT-1, remains to be determined. Nevertheless, identification of these epitopes should prove useful both for in vitro generation of potent tumoricidal WT-1–specific T cells for adoptive cell therapies and for the generation of more broadly applicable vaccines for stimulating T-cell responses for the eradication of clonogenic tumor cells expressing WT-1 in vivo.
There is an Inside Blood commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the National Institutes of Health (grants CA23766, CA59350, and CA08748), The Claire L. Tow Foundation, The Larry H. Smead Fund, The Aubrey Fund for Pediatric Cancer Research, The Ryan E. McGeough Charitable Gift Fund, The Major Family Fund for Cancer Research, and The Laura Rosenberg Foundation.
National Institutes of Health
Authorship
Contribution: E.D. and R.J.O. designed and conducted the study, analyzed the data, and wrote the manuscript; E.D., T.C., and D.P. conducted the experiments; and A.S. and A.H. provided the AAPCs, helped to design the validation experiments using AAPCs, and reviewed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard J. O'Reilly, MD, Department of Pediatrics and Medicine, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: oreillyr@mskcc.org.


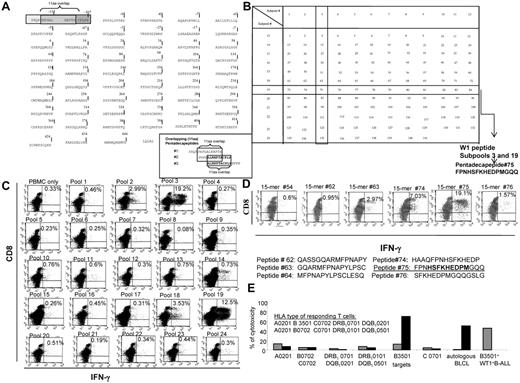
![Figure 3. HLA class I– and class II–restricted, WT-1–specific T cells respond to the same immunodominant peptide 15-mer derived from WT-1 protein in the WT-1 CTL sensitized with the WT-1 total pool of overlapping 15-mers loaded on autologous CAMs. (A) Production of IFNγ by the CD8+ and CD4+ WT-1–specific T cells in response to secondary overnight stimulation with the same dominant WT-1–derived 15-mer no. 41. (B) Identification of the immunogenic sequence of amino acids within pentadecapeptide no. 41 by IFNγ production after secondary overnight stimulation with autologous PBMCs loaded with a panel of 9-mers either unique for the peptide no. 41 (LDFAAPGAS [LDF]) or contained within the neighboring overlapping 15-mer no. 40 (PVLDFAPPG [PVL] or VLDFAPPGA [VLD]) and no. 42 (DFAPPGASA [DFA]). Only the 9-mer uniquely presented within the 15-mer no. 41, LDF, elicited an IFNγ response. (C) Peptide-specific cytotoxic activity of WT-1 CTLs against the panel of 9-mers and 11-mers contained within peptide no. 41 and loaded on autologous PHA-stimulated blasts is observed against both the 11-mer LDF and 9-mer LDF contained within the 11-mer LDF, as determined in a standard 51Cr-release assay at a 25:1 effector: stimulator ratio. (D) HLA restriction of the cytotoxic activity of the WT-1 CTLs. T cells restricted by HLA-A0201 lyse targets loaded with either the 11-mer or the 9-mer, whereas those restricted by HLA DRB10402 only lysed targets loaded with the 11-mer.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/120/8/10.1182_blood-2011-11-394619/4/m_zh89991293630003.jpeg?Expires=1769208859&Signature=Q1gcJDkAX1Ov5Eb~1jNz2DXpTGL7WdDmRYYJfZDyJzk98LL80CSz9KiAq4hrHjdqGCcYmvllrHHFNFQPfTmjZq2nExfFCxh2ed7YDE8wjWk~byjegqLCID2Ket4Ouywx4z-dFh5NPi~rhm680FkuWEr~b8CuQS4V5Lc5f7GSkqL~6k4QRmdstJXKfDYrrsFd82WGk5Aj~mgskE~bIpBpMAQoKMbIoLBrtEUCKcS2w2mZ51MU-lPmYVOmSlztonZAqnzB-y-cQpy0txSRFZjJmccj~gR-it~5~iwzCCbqNeDYxXSDUzyuZsZpUk5DbBbfkgf2s0KaRLwiAfJY590LOQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)