Abstract
Juvenile myelomonocytic leukemia (JMML) is a rare pediatric myeloid neoplasm characterized by excessive proliferation of myelomonocytic cells. Somatic mutations in genes involved in GM-CSF signal transduction, such as NRAS, KRAS, PTPN11, NF1, and CBL, have been identified in more than 70% of children with JMML. In the present study, we report 2 patients with somatic mosaicism for oncogenic NRAS mutations (G12D and G12S) associated with the development of JMML. The mutated allele frequencies quantified by pyrosequencing were various and ranged from 3%-50% in BM and other somatic cells (ie, buccal smear cells, hair bulbs, or nails). Both patients experienced spontaneous improvement of clinical symptoms and leukocytosis due to JMML without hematopoietic stem cell transplantation. These patients are the first reported to have somatic mosaicism for oncogenic NRAS mutations. The clinical course of these patients suggests that NRAS mosaicism may be associated with a mild disease phenotype in JMML.
Introduction
Juvenile myelomonocytic leukemia (JMML) is a rare myeloid neoplasm characterized by excessive proliferation of myelomonocytic cells. Somatic mutations in genes involved in GM-CSF signal transduction, such as NRAS, KRAS, PTPN11, NF1, and CBL, have been identified in more than 70% of children with JMML.1-3 The term “somatic mosaicism” is defined as the presence of multiple populations of cells with distinct genotypes in one person whose developmental lineages trace back to a single fertilized egg.4 Somatic mosaicism of various genes, including some oncogenes, has been implicated in many diseases. For example, somatic mosaicism for HRAS mutations is found in patients with Costello syndrome.5-7 Whereas germline mutations in causative genes (ie, PTPN11, NRAS, NF1, and CBL) are found in JMML patients,3,8-11 the presence of somatic mosaicism for these genes has never been reported. In the present study, we describe 2 cases of JMML in which the patients display somatic mosaicism for oncogenic NRAS mutations (G12D and G12S).
Study design
Written informed consent for sample collection was obtained from the patients' parents in accordance with the Declaration of Helsinki, and molecular analysis of the mutational status was approved by the ethics committee of the Nagoya University Graduate School of Medicine (Nagoya, Japan).
Patient 1.
A 10-month-old boy had hepatosplenomegaly and leukocytosis (72.1 × 109/L) with monocytosis (13.3 × 109/L; Table 1). The patient's BM contained 7% blasts with myeloid hyperplasia. Cytogenetic analysis revealed a normal karyotype and colony assay of BM mononuclear cells (BM-MNCs) showed spontaneous colony formation but GM-CSF hypersensitivity assay was not tested. The diagnostic criteria for JMML, as developed by the European Working Group on Myelodysplastic Syndrome in Childhood, was fulfilled,12 and the patient was treated with IFN-α and 6-mercaptopurine. His clinical and laboratory findings gradually resolved without hematopoietic stem cell transplantation. However, 11 years after the diagnosis of JMML, the patient developed thrombocytopenia (7.6 × 109/L) and BM findings showed trilineage dysplasia with low blast count compatible with refractory anemia. The patient did not have any physiologic abnormalities, such as facial deformity, and there was no family history of malignancy or congenital abnormalities.
Patient characteristics
| . | Patient 1 . | Patient 2 . |
|---|---|---|
| Age, mo | 10 | 10 |
| Sex | Male | Male |
| Liver, cm | 12 | 5 |
| Spleen, cm | 8 | 10 |
| WBCs, × 109/L | 72.1 | 31.8 |
| Monocytes, % | 18.5 | 20 |
| Blasts, % | 4 | 2 |
| Hb, g/dL | 8.9 | 5.4 |
| Platelets, × 109/L | 59 | 100 |
| HbF, % | 2.1 | 1.7 |
| BM blasts, % | 7 | 5 |
| Karyotype | 46,XY [20/20] | 46,XY [20/20] |
| Monosomy 7 (FISH) | Negative | Negative |
| Spontaneous colony formation | Positive | Positive |
| Gene mutation | NRAS, G12D 35G > A | NRAS, G12S 34G > A |
| Treatment | IFN-α-2b, 6-MP | None |
| Observation period, mo | 231 | 103 |
| Outcome | Alive | Alive |
| Fraction of mutant alleles, % (pyrosequencing) | ||
| Nail (whole) | 24 | 12.5 (average) |
| Nail (left hand) | ND | 26 |
| Nail (right hand) | ND | 13 |
| Nail (left foot) | ND | 8 |
| Nail (right foot) | ND | 3 |
| Buccal smear cells | 43 | 21 |
| Hair bulbs | 5 | ND |
| Family studies | ||
| Father | Wild-type | Wild-type |
| Mother | Wild-type | Wild-type |
| Sibling | ND | Wild-type |
| . | Patient 1 . | Patient 2 . |
|---|---|---|
| Age, mo | 10 | 10 |
| Sex | Male | Male |
| Liver, cm | 12 | 5 |
| Spleen, cm | 8 | 10 |
| WBCs, × 109/L | 72.1 | 31.8 |
| Monocytes, % | 18.5 | 20 |
| Blasts, % | 4 | 2 |
| Hb, g/dL | 8.9 | 5.4 |
| Platelets, × 109/L | 59 | 100 |
| HbF, % | 2.1 | 1.7 |
| BM blasts, % | 7 | 5 |
| Karyotype | 46,XY [20/20] | 46,XY [20/20] |
| Monosomy 7 (FISH) | Negative | Negative |
| Spontaneous colony formation | Positive | Positive |
| Gene mutation | NRAS, G12D 35G > A | NRAS, G12S 34G > A |
| Treatment | IFN-α-2b, 6-MP | None |
| Observation period, mo | 231 | 103 |
| Outcome | Alive | Alive |
| Fraction of mutant alleles, % (pyrosequencing) | ||
| Nail (whole) | 24 | 12.5 (average) |
| Nail (left hand) | ND | 26 |
| Nail (right hand) | ND | 13 |
| Nail (left foot) | ND | 8 |
| Nail (right foot) | ND | 3 |
| Buccal smear cells | 43 | 21 |
| Hair bulbs | 5 | ND |
| Family studies | ||
| Father | Wild-type | Wild-type |
| Mother | Wild-type | Wild-type |
| Sibling | ND | Wild-type |
Hb indicates hemoglobin; 6-MP, 6-mercaptopurine; and ND, not done.
Patient 2.
A 10-month-old boy had anemia, hepatosplenomegaly, and leukocytosis (31.8 × 109/L) with monocytosis (6.4 × 109/L; Table 1). The patient's BM exhibited myeloid hyperplasia and granulocytic dysplasia with 5% blasts. Cytogenetic analysis revealed a normal karyotype. Colony assay of BM-MNCs showed spontaneous colony formation and GM-CSF hypersensitivity. Although the diagnostic criteria for JMML were fulfilled,12 the patient's clinical symptoms and leukocytosis improved spontaneously within a few months without cytotoxic therapy or hematopoietic stem cell transplantation. The patient has remained healthy and has experienced no hematologic or physiologic abnormalities. The most recent follow-up examination was conducted when the patient was 8 years of age.
Detailed methods for experiments are described in supplemental Methods (available on the Blood Web site; see the Supplemental Materials link at the top of the online article).
Results and discussion
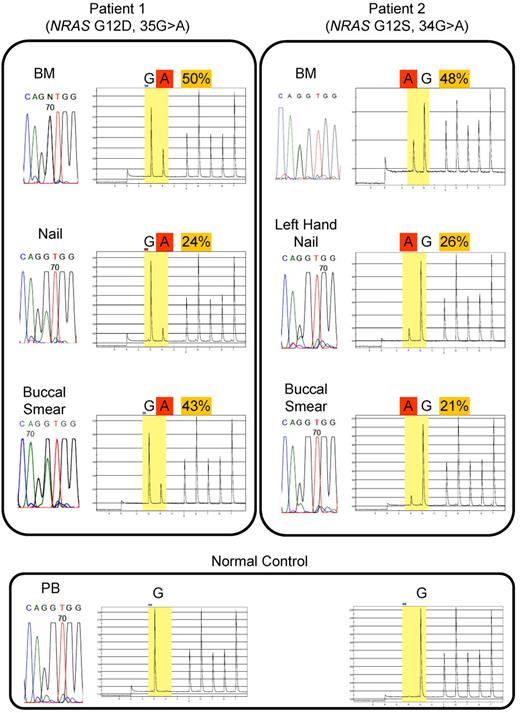
DNA sequencing for JMML-associated genes (ie, NRAS, KRAS, PTPN11, and CBL) was performed (Figure 1 and Table 1). In Patient 1, the NRAS G12D mutation was identified in BM-MNCs at the time of diagnosis of both JMML and MDS. We identified the same G12D mutation in DNA derived from buccal smear cells and nails of both hands; however, the sequence profile of the nails showed a low signal for the mutant allele compared with signal of blood cells. In Patient 2, the NRAS G12S mutation was identified in DNA from BM-MNCs, buccal smear cells, and nails of the left hand. However, the sequence profiles of buccal smear cells and nails of the left hand showed a low signal for the mutant variant. No mutation was detected in DNA from the PB-MNCs of the patient's parents or sibling.
Direct sequencing and quantitative mutational analysis of NRAS in JMML patients.NRAS mutations are detected by direct sequencing and quantified by pyrosequencing. Direct sequencing identified oncogenic NRAS mutations: for Patient 1, G12D, 35G > A; for Patient 2, G12S, 34G > A) in BM-MNCs at diagnosis of JMML and in the nails and buccal smear cells. Quantification by pyrosequencing revealed that the fractions of mutated allele varied among different tissue types. For Patient 1: BM, 50%; nail, 24%; and buccal smear, 43%. For Patient 2: BM, 48%; left-hand nail, 26%; and buccal smear, 21%.
Direct sequencing and quantitative mutational analysis of NRAS in JMML patients.NRAS mutations are detected by direct sequencing and quantified by pyrosequencing. Direct sequencing identified oncogenic NRAS mutations: for Patient 1, G12D, 35G > A; for Patient 2, G12S, 34G > A) in BM-MNCs at diagnosis of JMML and in the nails and buccal smear cells. Quantification by pyrosequencing revealed that the fractions of mutated allele varied among different tissue types. For Patient 1: BM, 50%; nail, 24%; and buccal smear, 43%. For Patient 2: BM, 48%; left-hand nail, 26%; and buccal smear, 21%.
We used pyrosequencing to quantify the fraction of mutated alleles in DNA samples from different somatic tissues (Figure 1 and Table 1). The frequency of mutated alleles varied by tissue type as follows. For Patient 1: BM-MNCs, 50%; nails, 24%; buccal smear cells, 43%; and hair bulbs, 5%. For Patient 2: buccal smear cells, 21%; nails of left hand, 26%; nails of right hand, 13%; nails of left foot, 8%; and nails of right foot, 3%. We cloned the PCR product of NRAS exon 2 from the nails of Patient 1 and picked up 15 clones. The clones were sequenced. Four of the 15 clones (27%) contained the mutant allele, which is consistent with the results of pyrosequencing analysis (24% mutant allele). Because the confirmed detection level by pyrosequencing technique was above 5%, results with a low percentage (< 5%) of mutant allele (ie, hair bulbs in Patient 1) should be interpreted with caution.13,14
We diagnosed 2 JMML patients as having somatic mosaicism of NRAS mutations: G12D for Patient 1 and G12S for Patient 2. The diagnoses were based on negative familial studies and mutational allele quantification analyses that showed diversity in the chimeric mutational status of different somatic tissues. Although DNA from buccal smear cells might be contaminated with WBCs, we also identified mutations in DNA from the nail tissue, which is known to be a good biologic material without contamination from hematopoietic cells, in both patients. These data suggest that a portion of the NRAS-mutated somatic cells were derived from one cell that acquired the mutation at a very early developmental stage. Although both somatic and germline mutations of RAS pathway genes (ie, PTPN11, NRAS, NF1, and CBL) are found in some JMML patients,3,8-11 somatic mosaicism for these genes has never been reported. To the best of our knowledge, the present study is the first report of JMML patients with somatic mosaicism of mutations in RAS pathway genes.
Germline RAS pathway mutations are often associated with dysmorphic features similar to Noonan syndrome or its associated diseases. Correspondingly, JMML patients with germline NRAS or CBL mutations exhibit characteristic dysmorphic features.3,10 Although our patients did not show any dysmorphic or developmental abnormalities, they should receive careful medical follow-up, especially for the occurrence of other cancers, because of the oncogenic nature of the mutations.
In general, JMML is a rapidly fatal disorder if left untreated.8 However, recent clinical genotype-phenotype analyses have revealed heterogeneity in their clinical course. We and other researchers have reported that patients with PTPN11 mutations have a worse prognosis than patients with other gene mutations, including NRAS and KRAS.15,16 Both of the JMML patients in the present study with somatic mosaicism of oncogenic NRAS mutations have had a mild and self-limiting clinical course. We analyzed nails of other 3 JMML patients with RAS mutations who experienced aggressive clinical course and none showed somatic mosaicism (data not shown). In analogy to the mild phenotype of JMML patients with germline mutations in PTPN11, we speculate that JMML patients with somatic mosaicism of RAS genes might have a mild clinical course. We are planning to confirm these observations in larger cohort.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ms Yoshie Miura, Ms Yuko Imanishi, and Ms Hiroe Namizaki for their valuable assistance with sample preparation and clerical work.
Authorship
Contribution: S.D. and H.M. designed and conducted the research, analyzed the data, and wrote the manuscript; A.S., M.M.-E., M. Sato, H.K., A.K., M. Sotomatsu, and Y.H. treated the patients; Y.T., Y.F.-H., K.Y., H.H., H.K., N.Y., H.S., A.N., X.W., O.I., Y.X., N.N., M.T., A.H., and K.K. conducted the research; and S.K. designed the research, analyzed the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Seiji Kojima, Department of Pediatrics, Nagoya University Graduate School of Medicine, 65 Tsuruma-cho, Showa-ku, Nagoya 466-8550, Japan; e-mail: kojimas@med.nagoya-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal