Abstract
Alternatively activated macrophages (AAMφ) are a major component of the response to helminth infection; however, their functions remain poorly defined. To better understand the helminth-induced AAMφ phenotype, we performed a systems-level analysis of in vivo derived AAMφ using an established mouse model. With next-generation RNA sequencing, we characterized the transcriptomes of peritoneal macrophages from BALB/c and IL4Rα−/− mice elicited by the nematode Brugia malayi, or via intraperitoneal thioglycollate injection. We defined expression profiles of AAMφ-associated cytokines, chemokines, and their receptors, providing evidence that AAMφ contribute toward recruitment and maintenance of eosinophilia. Pathway analysis highlighted complement as a potential AAMφ-effector function. Up-regulated mitochondrial genes support in vitro evidence associating mitochondrial metabolism with alternative activation. We mapped macrophage transcription start sites, defining over-represented cis-regulatory motifs within AAMφ-associated promoters. These included the binding site for PPAR transcription factors, which maintain mitochondrial metabolism. Surprisingly PPARγ, implicated in the maintenance of AAMφ, was down-regulated on infection. PPARδ expression, however, was maintained. To explain how PPAR-mediated transcriptional activation could be maintained, we used lipidomics to quantify AAMφ-derived eicosanoids, potential PPAR ligands. We identified the PPARδ ligand PGI2 as the most abundant AAMφ-derived eicosanoid and propose a PGI2-PPARδ axis maintains AAMφ during B malayi implantation.
Introduction
Macrophages display enormous functional diversity determined by signals from their immediate environment. IFNγ stimulation induces classic activation, an essential prerequisite for microbial infection control, whereas IL4/IL13 exposure polarizes macrophages toward alternative activation.1 Alternatively activated macrophages (AAMφ) are now implicated in the promotion of a wide range of diseases, including cancer,2 allergy,1 and fibrosis,1 but also in protection against helminth infection,3 diabetes,4 and obesity.4
Despite the flurry of interest in AAMφ, we remain remarkably ignorant of their physiologic role(s), partly because classic and alternative activation represents polar regions in a landscape of activation phenotypes sculpted by multiple factors. Different cellular developmental histories,5 microenvironmental cues, and factor-dependent polarization6 vastly increase the complexity of macrophage phenotypes in vivo. IL4/IL13 induce canonical alternative activation via IL4Rα-dependent phosphorylation of STAT6, driving the transcription of a diverse repertoire of genes, including Arg1 (Arginase-1), Chi3l3 (Chitinase 3-like 3, YM-1), and Retnla (resistin-like α, RELMα, FIZZ-1). Because exposure to helminths almost universally induces potent Th2 responses, alternative activation of Mφ is characteristic of these infections.7 Indeed, Chi3l3 and Retnla were described as AAMφ markers associated with challenge by the parasitic nematode Brugia malayi.7
An emerging paradigm suggests that cytokine-mediated alterations in cellular metabolism determine cellular life-history8 and effector functions.9 For example, a switch from glucose dependency to mitochondrial metabolism oversees the ability of effector CD8+ T cells to commit to a memory phenotype.8 In this context, it is interesting to note that in vitro studies suggest that classically activated macrophages (CAMφ) and AAMφ are associated with different metabolic profiles. CAMφ require aerobic glycolysis,10 whereas AAMφ couple lipid oxidation with oxidative phosphorylation.4 Cooperative interactions between STAT6, PPARγ, and PGC-1β are considered necessary4,11 to induce these metabolic changes in AAMφ. In vivo confirmation of this observation is required, and an improved understanding of the role for mitochondrial metabolism in alternative activation may yield key insights into the effector functions of these cells.
Previous transcriptome analyses of AAMφ have used in vitro-generated cells, whereas in vivo studies of Th2 environments have analyzed whole tissue.12,13 This leaves a gap in our understanding of AAMφ function during infection. Previously, we identified abundantly expressed genes in in vivo–derived AAMφ using an expressed sequence tag approach.7 This provided valuable insight into markers expressed by these cells but lacked the power to critically assess molecular pathways associated with alternative activation in vivo. Here, using second-generation Illumina sequencing and mass spectrometry, we combined transcriptomics and lipidomics to gain a global overview of macrophage IL4Rα-dependent transcription and transcriptional regulation in vivo.
We compared nematode-elicited macrophages (NeMφ) and inflammatory-like thioglycollate-elicited macrophages (ThioMφ) from both wild-type (WT) and IL4Rα−/− mice. We sought to define physiologic functions of in vivo–derived AAMφ and have focused on understanding the macrophage response to filarial nematode infection. We defined macrophages as F4/80-positive cells with a negative gating strategy to exclude contaminants. This definition probably includes macrophage subpopulations that contribute differentially toward the overall response; however, this comparison allowed us to focus on the most relevant changes in macrophage physiology to filarial nematode challenge. The contrast between WT-NeMφ and WT-ThioMφ identified differential gene expression due to the presence of the nematode, or differences in cell origin. Comparing WT-NeMφ and IL4Rα−/−-NeMφ revealed IL4Rα-dependent components of macrophage activation during helminth infection. We followed Siamon Gordon's definition of alternative activation as the IL4/IL13–dependent component of macrophage activation.1 IL4Rα deficiency ablates both IL4– and IL13–dependent signaling.1 Thus, coordinately differentially expressed (DE) genes in WT-NeMφ (ie, in vivo generated AAMφ) relative to both WT-ThioMφ and IL4Rα−/−-NeMφ are, by definition, those relevant to alternative activation during helminth infection.
Illumina RNA sequencing (RNA-Seq) provided > 5 orders of magnitude dynamic range between the most abundant and lowly expressed genes, delivering the most extensive characterization of in vivo–polarized macrophage populations to date. We establish a putative role for AAMφ in eosinophil recruitment and the complement response during helminth infection. Macrophage transcription start sites (TSSs) were mapped; and, by characterizing over-represented cis-regulatory elements in AAMφ promoters, we confirm PPAR-dependent transcription as a major facilitator of alternative activation in vivo. Pathway analysis supported these findings by identifying AAMφ-dependent alterations in lipid and mitochondrial metabolism, key targets of PPAR transcription factors. Finally, liquid chromatography-tandem mass spectrometry (LC-MS/MS) was used to define the B malayi–induced repertoire of endogenous eicosanoids, allowing us to propose a mechanism for PPARδ-mediated alternative activation. We thus provide global mechanistic insights into the function and regulation of helminth-elicited AAMφ and identify putative effector molecules involved in the maintenance and regulation of alternative activation in vivo.
Methods
Generation of macrophage populations
Nine WT BALB/c and 9 IL4Rα−/− mice were infected with 4 B malayi adult females and 1 male by surgical implant, or challenged with an intraperitoneal thioglycollate injection (700 μL of 4% Brewer-modified thioglycollate medium in PBS [weight/volume], BD Biosciences Pharmingen) as described previously.5 All work was conducted in accordance with the Animals (Scientific Procedures) Act of 1986. B malayi were obtained from Mongolian jirds purchased from TRS Laboratories. Twenty-one days after implantation, or 3 days after thioglycollate treatment, whole peritoneal exudate cells (PECs) were extracted by lavage with RPMI + HEPES, 1% penicillin/streptomycin, 2mM EDTA, and macrophages purified by FACS.
FACS purification and intracellular cytokine staining
PECs were treated with red blood cell lysis media (Sigma-Aldrich) and up to 1 × 107 cells retained per mouse for FACS sorting. PEC cells were stained with Live/Dead Aqua (Invitrogen), F4/80-biotin (BioLegend), SiglecF-PE, B220-PE, CD4-FITC, and streptavidin-allophycocyanin (eBioscience) before sorting on a FACSAria (BD Biosciences). F4/80+ macrophages were sorted based on allophycocyanin positivity. Negative gating based on PE and FITC staining was used to ensure the highest possible purity (Figure 1; supplemental Figure 1, see the Supplemental Materials link at the top of the article). Macrophage purity was verified by flow cytometry and stored in Qiazol (QIAGEN) before RNA extraction. Alternative activation in WT-infected mice was confirmed using intracellular staining for RELMα as described previously.5
Flow cytometric acquisition of macrophages and confirmation of alternative activation in WT B malayi–implanted macrophages. (A) Gating strategy used to obtain pure macrophage populations as shown with 1 representative WT B malayi–infected individual. After the removal of dead cells, B220, SiglecF, and CD4-positive cells were excluded. F4/80 high cells were then selected and doublets removed based on forward scatter width (FSC-W)/forward scatter area (FSC-A). (B) Intracellular cytokine staining for RELMα expression in thioglycollate-elicited macrophages (top) and B malayi–elicited macrophages (bottom). The scatter profile and macrophage gates on the left refer to 1 representative WT B malayi–infected individual. (C) Bar chart showing percentage of RELMα-positive macrophages from analysis. (B) n = 9 per group. ***P < .001.
Flow cytometric acquisition of macrophages and confirmation of alternative activation in WT B malayi–implanted macrophages. (A) Gating strategy used to obtain pure macrophage populations as shown with 1 representative WT B malayi–infected individual. After the removal of dead cells, B220, SiglecF, and CD4-positive cells were excluded. F4/80 high cells were then selected and doublets removed based on forward scatter width (FSC-W)/forward scatter area (FSC-A). (B) Intracellular cytokine staining for RELMα expression in thioglycollate-elicited macrophages (top) and B malayi–elicited macrophages (bottom). The scatter profile and macrophage gates on the left refer to 1 representative WT B malayi–infected individual. (C) Bar chart showing percentage of RELMα-positive macrophages from analysis. (B) n = 9 per group. ***P < .001.
RNA-Seq library preparations, high-throughput sequencing, and bioinformatic analyses
Full details are provided in the supplemental Methods. Briefly, RNA was extracted using the QIAGEN miRNeasy kit and quality assessed on an Agilent Bioanalyzer. RNA-Seq libraries were prepared from 2 μg of RNA, each obtained by pooling 0.66 μg from 3 mice, using the Illumina paired-end RNA-Seq library preparation kit. Fifty-one base paired-end RNA-Seq libraries were sequenced on an Illumina GAIIx, and reads were mapped to the mouse reference genome using TopHat. Differential expression analysis was performed using DESeq, and TSSs predicted using the custom algorithm TSS-Predictor. Gene set enrichment and hierarchical cluster analysis were performed in the R environment.
Lipidomics
Full details of the lipidomic experimental methods are given in supplemental Methods. Briefly, in an independent homologous experiment to the RNA-Seq, adherence purified macrophages from whole PECs were cultured for 12 hours. Total eicosanoids were extracted from peritoneal lavage supernatant and purified macrophage cultures using C18 solid phase extraction cartridges, after spiking with known amounts of the deuterated internal standards 15-hydroxyeicosatetraenoic acid (15-HETE)-d8, LTB4-d4, and PGE2-d4. Eicosanoids were separated on a Thermo Scientific Hypersil Gold C18 column (3 μm × 2.1 × 50 mm) directed into an online tandem mass spectrometer (TSQ Quantum Ultra; Thermo Scientific) operating in negative ion mode. Data were acquired and analyzed using LCquan Version 2.6 software (Thermo Scientific).
Results
Generation of AAMφ and differential expression analysis
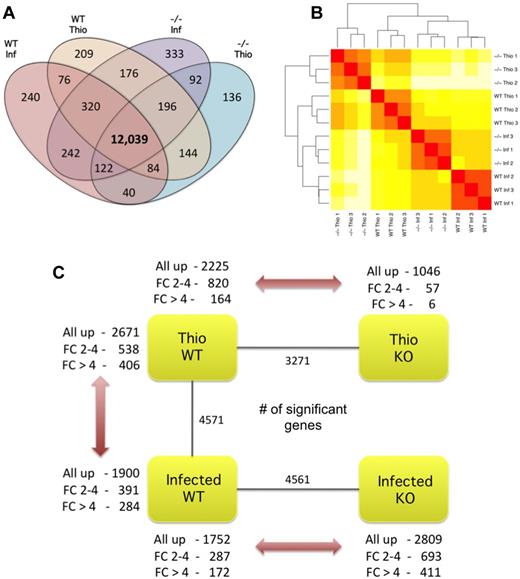
We generated NeMφ and ThioMφ by implanting BALB/c and IL4Rα−/− mice with the nematode B malayi, or via intraperitoneal administration of thioglycollate to elicit a population of nonpolarized, inflammatory-like macrophages. IL4 stimulation increases F4/80 surface expression on macrophages.5 To enrich for AAMφ in the implant setting, we collected the brightest F4/80+ macrophage population in each condition (supplemental Figure 1). Macrophage purity was maximized by exclusion of dead cells, doublets, B cells (B220+), eosinophils (SiglecF+), and CD4+ T cells using negative gating (Figure 1A). Our sorting strategy resulted in an average of 97.3% purity (range, 94.9%-99.4%, supplemental Table 1). Alternative activation of WT-NeMφ was confirmed with intracellular cytokine staining for RELMα (Figure 1B-C). On average, 47% of WT-NeMφ were RELMα positive, consistent with 30%-60% positivity typically seen after B malayi implant. RNA-Seq libraries yielded between 11 million and 30 million 51-base paired-end reads. Gene expression was quantified by mapping reads to the mouse reference genome using TopHat.14 In total 55%-73% of reads mapped uniquely to the genome, with 7.7-25 million mapping within exons of known genes (Ensembl Version 58, Table 1). Between 12 853 and 13 520 (56%-59%) protein coding genes were expressed in each group, with 12 039 of these common to all 4 populations (Figure 2A). We validated sample purity by assessing the expression of lineage-restricted marker genes for potential contaminants; eosinophils, neutrophils, and B cells (supplemental Table 2). With this approach, we confirmed negligible contamination of neutrophils or eosinophils; however, a low level of Cd19 expression was observed in IL4Rα−/−-NeMφ.
Sequence and mapping statistics for raw Illumina data
| Strain . | Treatment . | Repeat . | Total reads . | No. mapped . | % mapped . | Properly paired . | % properly paired . | Singletons . | % singletons . | Unique reads in ensembl v58 annotations . | % unique reads in ensembl v58 annotations . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BALB/c | Infected | 1 | 35 646 760 | 24 026 742 | 67.4 | 18 883 064 | 78.59 | 3 802 882 | 15.83 | 13 914 673 | 57.9 |
| BALB/c | Infected | 2 | 39 663 694 | 26 450 844 | 73.3 | 21 349 672 | 80.71 | 4 071 578 | 15.39 | 15 261 028 | 57.7 |
| BALB/c | Infected | 3 | 40 960 176 | 26 670 826 | 65.1 | 20 637 676 | 77.38 | 4 486 602 | 16.82 | 15 578 573 | 58.4 |
| BALB/c | Thio | 1 | 40 398 232 | 26 011 652 | 64.3 | 21 817 520 | 83.88 | 3 454 426 | 13.28 | 14 732 920 | 56.6 |
| BALB/c | Thio | 2 | 48 366 774 | 33 194 643 | 68.6 | 26 782 502 | 80.68 | 5 494 173 | 16.55 | 19 344 275 | 58.2 |
| BALB/c | Thio | 3 | 45 552 296 | 28 796 129 | 63.2 | 24 350 132 | 84.56 | 4 167 411 | 14.47 | 16 480 896 | 57.2 |
| IL4Rα−/− | Infected | 1 | 56 700 552 | 31 428 896 | 55.4 | 27 125 044 | 86.31 | 4 172 298 | 13.28 | 17 799 754 | 56.6 |
| IL4Rα−/− | Infected | 2 | 23 150 358 | 15 076 797 | 65.1 | 12 977 824 | 86.08 | 1 911 075 | 12.68 | 8 493 659 | 56.3 |
| IL4Rα−/− | Infected | 3 | 60 376 960 | 42 872 365 | 71.0 | 32 888 162 | 76.71 | 6 826 787 | 15.92 | 24 849 426 | 57.9 |
| IL4Rα−/− | Thio | 1 | 40 492 838 | 24 144 541 | 59.6 | 19 772 782 | 81.89 | 3 547 653 | 14.69 | 13 845 966 | 57.3 |
| IL4Rα−/− | Thio | 2 | 22 996 482 | 13 033 134 | 56.7 | 8 952 742 | 68.69 | 2 418 168 | 18.55 | 7 725 619 | 59.2 |
| IL4Rα−/− | Thio | 3 | 36 062 844 | 19 911 073 | 55.2 | 14 751 228 | 74.09 | 3 874 775 | 19.46 | 11 892 859 | 59.7 |
| Strain . | Treatment . | Repeat . | Total reads . | No. mapped . | % mapped . | Properly paired . | % properly paired . | Singletons . | % singletons . | Unique reads in ensembl v58 annotations . | % unique reads in ensembl v58 annotations . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BALB/c | Infected | 1 | 35 646 760 | 24 026 742 | 67.4 | 18 883 064 | 78.59 | 3 802 882 | 15.83 | 13 914 673 | 57.9 |
| BALB/c | Infected | 2 | 39 663 694 | 26 450 844 | 73.3 | 21 349 672 | 80.71 | 4 071 578 | 15.39 | 15 261 028 | 57.7 |
| BALB/c | Infected | 3 | 40 960 176 | 26 670 826 | 65.1 | 20 637 676 | 77.38 | 4 486 602 | 16.82 | 15 578 573 | 58.4 |
| BALB/c | Thio | 1 | 40 398 232 | 26 011 652 | 64.3 | 21 817 520 | 83.88 | 3 454 426 | 13.28 | 14 732 920 | 56.6 |
| BALB/c | Thio | 2 | 48 366 774 | 33 194 643 | 68.6 | 26 782 502 | 80.68 | 5 494 173 | 16.55 | 19 344 275 | 58.2 |
| BALB/c | Thio | 3 | 45 552 296 | 28 796 129 | 63.2 | 24 350 132 | 84.56 | 4 167 411 | 14.47 | 16 480 896 | 57.2 |
| IL4Rα−/− | Infected | 1 | 56 700 552 | 31 428 896 | 55.4 | 27 125 044 | 86.31 | 4 172 298 | 13.28 | 17 799 754 | 56.6 |
| IL4Rα−/− | Infected | 2 | 23 150 358 | 15 076 797 | 65.1 | 12 977 824 | 86.08 | 1 911 075 | 12.68 | 8 493 659 | 56.3 |
| IL4Rα−/− | Infected | 3 | 60 376 960 | 42 872 365 | 71.0 | 32 888 162 | 76.71 | 6 826 787 | 15.92 | 24 849 426 | 57.9 |
| IL4Rα−/− | Thio | 1 | 40 492 838 | 24 144 541 | 59.6 | 19 772 782 | 81.89 | 3 547 653 | 14.69 | 13 845 966 | 57.3 |
| IL4Rα−/− | Thio | 2 | 22 996 482 | 13 033 134 | 56.7 | 8 952 742 | 68.69 | 2 418 168 | 18.55 | 7 725 619 | 59.2 |
| IL4Rα−/− | Thio | 3 | 36 062 844 | 19 911 073 | 55.2 | 14 751 228 | 74.09 | 3 874 775 | 19.46 | 11 892 859 | 59.7 |
Overall gene expression and differential expression analysis. (A) Number of expressed genes in macrophage populations where expressed is considered as at least 1 read mapping to a gene in all 3 replicates of a condition. (B) Unsupervised, hierarchical clustering of individual lanes demonstrating discrete clustering of biologic replicates. (C) Summary of DE genes (P < .01) in each pairwise comparison showing the total number of DE genes (inner), and a breakdown showing the direction of differential expression for both moderately (log 2-fold change ± 2-4) and highly (log 2-fold change > 4) DE genes.
Overall gene expression and differential expression analysis. (A) Number of expressed genes in macrophage populations where expressed is considered as at least 1 read mapping to a gene in all 3 replicates of a condition. (B) Unsupervised, hierarchical clustering of individual lanes demonstrating discrete clustering of biologic replicates. (C) Summary of DE genes (P < .01) in each pairwise comparison showing the total number of DE genes (inner), and a breakdown showing the direction of differential expression for both moderately (log 2-fold change ± 2-4) and highly (log 2-fold change > 4) DE genes.
Hierarchical clustering of global gene expression profiles grouped RNA-Seq libraries according to biologic condition, reaffirming the quality and reproducibility of our analysis (Figure 2B). After differential expression analysis, we identified substantial transcriptional differences between the macrophage populations in the 3 key comparisons (Figure 2C). Using a P value cutoff of .01 after correction for multiple testing (Benjamini-Hochberg method), we identified 4571 DE genes between WT-NeMφ and WT-ThioMφ, 4561 DE genes between WT-NeMφ and IL4Rα−/−-NeMφ, and 3271 DE genes between WT-ThioMφ and IL4Rα−/−-ThioMφ (Figure 2C). A complete gene list and associated P values are provided in supplemental Table 3. We observed a higher number of DE genes between WT-NeMφ and IL4Rα−/−-NeMφ than between WT- ThioMφ and IL4Rα−/−-ThioMφ (Figure 2C). In addition, the magnitudes of these differences were much greater in the infection setting (supplemental Figure 7). Furthermore, a similar range of differential expression was observed between WT-NeMφ and WT-ThioMφ as for WT-NeMφ and IL4Rα−/−-NeMφ. Thus, as expected, IL4Rα-dependent signaling drove major alterations in the macrophage transcriptional profile in response to Th2-inducing immune stimuli.
We interrogated the expression profiles of DE genes to identify those explicitly associated with alternative activation. All DE genes from the WT-NeMφ versus WT-ThioMφ and WT-NeMφ versus IL4Rα−/−-NeMφ comparisons (4571 and 4561, respectively) were grouped according to their expression profile using hierarchical agglomerative clustering. The resulting tree was subdivided into 20 clusters, and the expression profile of genes in each cluster was assessed (supplemental Figure 8). We identified 5 clusters with increased expression levels in WT-NeMφ relative to both WT-ThioMφ and IL4Rα−/−-NeMφ. These were classified as AAMφ-up (Figure 3; supplemental Table 4). Similarly, 5 clusters were identified with converse expression profiles and classified as AAMφ-down. In total, 1658 genes AAMφ-up genes and 1735 AAMφ-down genes were identified (Figure 3). Importantly, we can state that expression of genes in AAMφ-up and AAMφ-down clusters is IL4Rα-dependent. However, as the IL4Rα−/− mice lack IL4Rα expression on all cells, we cannot say whether these effects are cell-autonomous and, in some cases, may reflect other changes, such as Th2 cell activation.
Expression profiles of differentially expressed AAMφ-associated gene sets. The expression profiles of gene sets positively and negatively associated with alternative activation (AAMφ-up and AAMφ-down). All statistically significant genes (P < .01) between WT-NeMφ and WT-ThioMφ, and WT-NeMφ and IL4Rα−/−-NeMφ, were clustered using hierarchical agglomerative clustering. Each gene within an expression cluster is plotted in blue, and the mean expression for all genes within each cluster is overlaid in black. The figure in each panel represents the total number of genes in that cluster.
Expression profiles of differentially expressed AAMφ-associated gene sets. The expression profiles of gene sets positively and negatively associated with alternative activation (AAMφ-up and AAMφ-down). All statistically significant genes (P < .01) between WT-NeMφ and WT-ThioMφ, and WT-NeMφ and IL4Rα−/−-NeMφ, were clustered using hierarchical agglomerative clustering. Each gene within an expression cluster is plotted in blue, and the mean expression for all genes within each cluster is overlaid in black. The figure in each panel represents the total number of genes in that cluster.
Differential expression of immune effectors
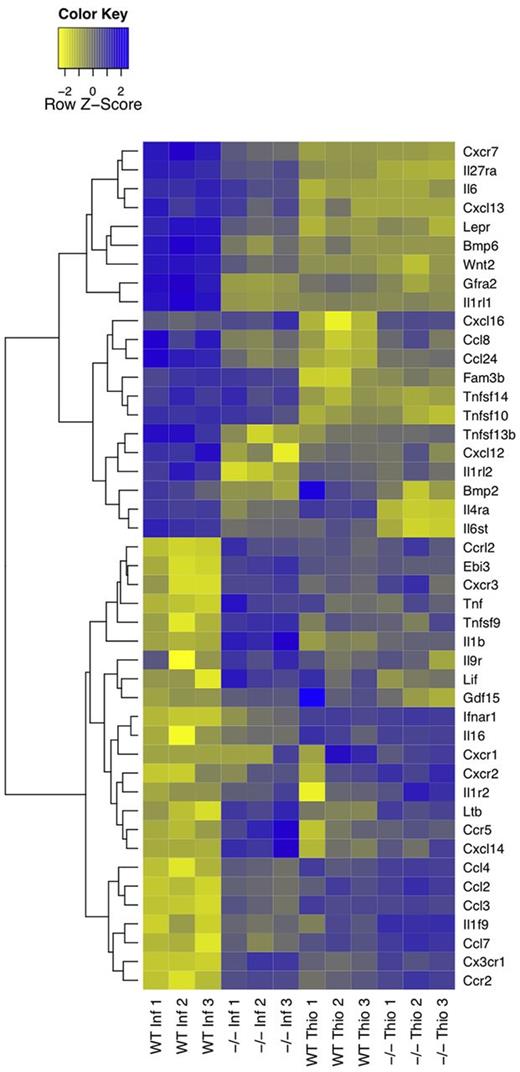
We reasoned that a descriptive, knowledge-based, assessment of immunologically relevant GO terms (encompassing cytokines, chemokines, and their receptors; GO terms GO:0005125, GO:0008009, GO:0004896, and GO:0004950, respectively; hereafter called immune effectors) would provide insight into AAMφ function and regulation. Immune effectors that were present in AAMφ-up and AAMφ-down clusters were defined as AAMφ-associated. Using this criterion, we identified AAMφ-association for 16 cytokines, 11 cytokine receptors, 10 chemokines, and 8 chemokine receptors (Figure 4). Here we discuss the function of key AAMφ-associated immune effectors, providing insight into the IL4Rα-dependent facets of the macrophage response to filarial nematode infection in vivo and defining candidate genes for future investigations.
Heatmap of alternative activation-modulated immune effectors. Immune effector genes, cytokines, chemokines, and their respective receptors (GO:0005125, GO:0008009, GO:0004896, and GO:0004950, respectively), in AAMφ-associated clusters were identified based on Gene Ontology annotations. Hierarchical clustering analysis reveals a unique expression profile of AAMφ-associated immune effectors.
Heatmap of alternative activation-modulated immune effectors. Immune effector genes, cytokines, chemokines, and their respective receptors (GO:0005125, GO:0008009, GO:0004896, and GO:0004950, respectively), in AAMφ-associated clusters were identified based on Gene Ontology annotations. Hierarchical clustering analysis reveals a unique expression profile of AAMφ-associated immune effectors.
Chemokine and chemokine receptor expression profiles.
The AAMφ chemokine expression profile (Figure 4) was consistent with macrophage-mediated maintenance of the cellular milieu in the peritoneal cavity of B malayi–implanted mice, which is composed primarily of B cells, macrophages, and eosinophils.15 All assayed macrophage populations abundantly and constitutively expressed Ccl6 and Ccl9 (supplemental Table 5), which may serve to maintain macrophage populations within the peritoneal cavity.16 In the AAMφ-up clusters, we identified the eosinophil chemoattractants Ccl24 and Ccl8,16 as abundantly and moderately expressed, respectively. Cxcl12 and Cxcl13 were also identified as AAMφ-associated, however, were expressed at much lower levels. Consistent with an anti-inflammatory or noninflammatory phenotype of AAMφ,1 the AAMφ-down clusters contained Ccl3 (Mip1α), Ccl4, Ccl2 (MCP-1), Ccl7 (MCP-3), and Cxcl14, all of which are involved in acute phase inflammation, attracting primarily monocytes and neutrophils.17
Interestingly, AAMφ-up clusters were devoid of chemokine receptors with the exception of Cxcr7 (Figure 4). Cxcr7 responds to the stromal-derived Cxcl12, affecting both migration and differentiation in monocytes.18 Ccr1 (MIP-1α receptor, supplemental Table 5) was also highly expressed by macrophages, but not classified as AAMφ-associated. Therefore, Cxcr7 and/or Ccr1 may be key determinants of nematode-induced AAMφ localization.17,18 A large number of chemokine receptors were present in the AAMφ-down clusters. These included Ccr2, Cxcr1, Cxcr2, Cxcr3, Ccr5, and Ccrl2, implying that AAMφ are impaired in their capacity to respond to numerous chemokines. This suggests that WT-NeMφ do not migrate to prime lymphatic T-cell responses, despite high MHCII expression and functional antigen presentation.19,20
Cytokine and cytokine receptor expression profiles.
Cytokine receptor expression was typically maintained or enhanced in NeMφ. AAMφ-up clusters contained the IL33 receptor Il1rl1 (ST-2) and the IL27 receptor subunit Il27ra (WSX-1), both of which have previously been characterized as AAMφ-associated.21,22 AAMφ-up clusters also included Gfra2 (GDNF family receptor α-2), involved in neuronal survival and differentiation,23 and the leptin receptor (Lepr). Leptin is an adipokine involved in energy homeostasis with broad, pleiotropic effects. There is no described role for leptin in Th2 immunity; however, macrophages of Lepr-deficient mice express higher levels of inflammatory cytokines,24 suggesting the possibility that the leptin receptor contributes to the anti-inflammatory profile of AAMφ.
With regard to cytokines, WT-NeMφ produced significantly more Il6, Wnt2, and Bmp6 than either WT-ThioMφ or IL4Rα−/−-NeMφ. Bmp6 is pleiotropic, suppressing B-cell proliferation25 and promoting macrophage IL6 production.26 Wnt2 has no defined role in macrophage physiology, although Wnts do influence immune cell fate decisions.27 Thus, Wnt2 and Bmp6 represent promising novel candidates for future investigations of Th2 immunity.
AAMφ are described as broadly anti-inflammatory,1 in part because of their production of IL10. Surprisingly, in the chronic setting of our experiment, WT-NeMφ did not produce any IL10 (supplemental Table 5). Nonetheless, AAMφ-down clusters contained many proinflammatory cytokines. Among these were Tnf, Lif, Il1b, and Il16.28-30 In addition, the expression of the macrophage activating factor Tnfsf931 and the IL27 and IL35 subunit Ebi3 were lowered in AAMφ. In summary, AAMφ expressed greater levels of immune effectors associated with eosinophil recruitment and survival and lower levels of numerous proinflammatory agents.
KEGG pathway analysis
To assess whether metabolic changes in macrophages previously observed during IL4 treatment in vitro also occur in vivo, we applied gene set enrichment analysis (GSEA) using the KEGG pathway database alongside modifications for RNA-Seq data (see supplemental Methods). By considering the relative expression of all genes, rather than only DE genes, GSEA provides a sensitive metric for identifying differences in biochemical and cell signaling pathways.
The complement and coagulation cascade.
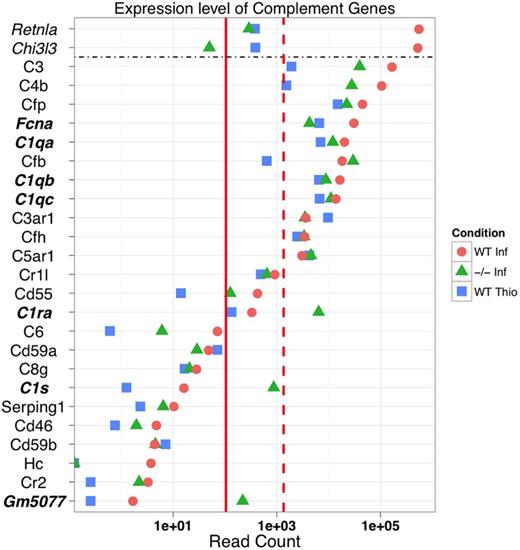
Unexpectedly, the complement and coagulation cascade (KEGG pathway mmu04610) was the most differentially regulated pathway between WT-NeMφ and WT-ThioMφ (supplemental Table 6). The most striking feature of complement expression in AAMφ was transcript abundance (Figure 5). For example, C3 and C4 attracted > 100 000 reads each, making them among the most abundant NeMφ-associated transcripts. NeMφ showed up-regulation of C1q complex genes (C1qa, C1qb, and C1qc), and striking overexpression of MBL pathway constituents, specifically FicolinA (FcnA), Cfb, and Cfp (supplemental Figure 9). This suggests a role for FicolinA-dependent complement activity in the response to helminth infection.
Complement components are abundantly expressed in an alternative activation-dependent manner. The expression of genes in the complement and coagulation cascade (KEGG pathway mmu:04610). For reference, the median and 90th percentiles of expression for all expressed genes are included (solid and dashed red lines, respectively). In addition, highly expressed marker genes Chi3l3 (YM-1) and Retnla (RELMα) are included for reference.
Complement components are abundantly expressed in an alternative activation-dependent manner. The expression of genes in the complement and coagulation cascade (KEGG pathway mmu:04610). For reference, the median and 90th percentiles of expression for all expressed genes are included (solid and dashed red lines, respectively). In addition, highly expressed marker genes Chi3l3 (YM-1) and Retnla (RELMα) are included for reference.
Mitochondrial metabolism.
Twenty-three of the 27 most differentially regulated pathways between WT-NeMφ and WT-ThioMφ were metabolic (supplemental Figure 10; supplemental Table 6). A similar trend was observed between WT-NeMφ and IL4Rα−/−-NeMφ, supporting a model wherein a shift in metabolic phenotype is a cardinal feature of alternative activation4 (supplemental Figure 10). The second most perturbed KEGG pathway between WT-NeMφ and WT-ThioMφ was the tricarboxylic acid (TCA) cycle (supplemental Table 6). Indeed, the majority of expressed TCA cycle genes were expressed at a higher level in WT-NeMφ relative to both WT-ThioMφ and IL4Rα−/−-NeMφ (Figure 6A; supplemental Figure 11). A similar pattern of gene expression was also observed for genes involved in oxidative phosphorylation (Figure 6B). Taken together, these findings are consistent with previous studies showing that IL4 induces expansion of the mitochondrial compartment in macrophages in vitro.32
AAMφ up-regulate mitochondrial TCA cycle genes and show evidence of PPAR-dependent transcription in vivo. (A) Schematic of the TCA cycle showing DE genes in WT-NeMφ relative to WT-ThioMφ. Blue border represents up-regulated; and yellow border, down-regulated. In addition, a red asterisk indicates genes up-regulated in WT-NeMφ relative to IL4Rα−/−-NeMφ. (B) Heatmap showing relative expression of mitochondrial electron transport chain (ETC) components between WT-NeMφ, IL4Rα−/−-NeMφ, and WT-ThioMφ. The majority of ETC components are most highly expressed in WT-NeMφ. (C) The expression levels of transcripts encoding for PPARγ, PPARδ, and the coactivator protein PGC-1β show that WT-NeMφ express high levels of PPARδ and PGC-1β, but not PPARγ. (D) Confirmation of the accuracy of TSS prediction. The density plot shows the distances from the predicted TSS for each gene to the nearest annoated Ensembl TSS (median absolute deviation = 32 bp). (E) Consensus motifs for the 3 over-represented transcription factor binding sites in AAMφ-associated promoter regions identified using Clover (P < .01), comparison of AAMφ-associated promoter regions (TSS −400, +100 bp) against non-AAMφ-associated macrophage promoters.
AAMφ up-regulate mitochondrial TCA cycle genes and show evidence of PPAR-dependent transcription in vivo. (A) Schematic of the TCA cycle showing DE genes in WT-NeMφ relative to WT-ThioMφ. Blue border represents up-regulated; and yellow border, down-regulated. In addition, a red asterisk indicates genes up-regulated in WT-NeMφ relative to IL4Rα−/−-NeMφ. (B) Heatmap showing relative expression of mitochondrial electron transport chain (ETC) components between WT-NeMφ, IL4Rα−/−-NeMφ, and WT-ThioMφ. The majority of ETC components are most highly expressed in WT-NeMφ. (C) The expression levels of transcripts encoding for PPARγ, PPARδ, and the coactivator protein PGC-1β show that WT-NeMφ express high levels of PPARδ and PGC-1β, but not PPARγ. (D) Confirmation of the accuracy of TSS prediction. The density plot shows the distances from the predicted TSS for each gene to the nearest annoated Ensembl TSS (median absolute deviation = 32 bp). (E) Consensus motifs for the 3 over-represented transcription factor binding sites in AAMφ-associated promoter regions identified using Clover (P < .01), comparison of AAMφ-associated promoter regions (TSS −400, +100 bp) against non-AAMφ-associated macrophage promoters.
PPARγ and PGC-1β, key regulators of mitochondrial metabolism, have been described as required for alternative macrophage activation in vitro.12,32 Accordingly, the metabolic profile we observe could be explained by transcriptional activity of PPAR family members (PPARα, PPARγ, and PPARδ). However, during B malayi infection, macrophage-specific PPARγ−/− mice show no impairment in alternative activation (D.R., unpublished data, October 2009). Furthermore, although PGC-1β expression was augmented in an IL4Rα- and infection-dependent manner, PPARγ levels were substantially lower in NeMφ than ThioMφ (Figure 6C). PPARα was not expressed by any of the assayed macrophage populations (supplemental Table 5). Only PPARδ expression was sustained in NeMφ, yet it was not up-regulated (Figure 6C). This suggests that, during helminth infection, PPARδ may compensate, either partially, or completely, for reduced PPARγ in NeMφ. Alternatively, PPAR-independent transcription may maintain mitochondrial metabolism in NeMφ.
TSS identification and promoter region analysis
To address whether PPARs facilitate transcription of IL4Rα-dependent genes in vivo we analyzed transcription factor binding sites (TFBSs) in the promoters of AAMφ-associated genes. We developed and optimized a method to accurately identify TSSs using RNA-Seq data (Figure 6D; for details of the method and validation, see supplemental Methods). Briefly, transcripts were defined using Cufflinks,33 and TSSs identified based on expected gene expression and the observed sequencing read depth at each position in the gene. Proximal promoter sequences (300 bp upstream and 100 bp downstream of each TSS) were obtained from 7817 high-confidence TSSs and analyzed for over-represented TFBSs.
We validated our method by identifying over-represented TFBSs in our promoters relative to all mouse promoters using Clover,34 and comparing the found set with the macrophage lineage-restricted TFBSs identified by Hume et al.35 In our data, we identified 54 motifs that were present significantly more often than expected (P < .01). Hume et al35 used a different library of TFBS motifs and identified 27 significant motifs, 23 of which were represented in the library we used. Our analysis identified 18 of these (78%) as over-represented in our macrophage promoters, reinforcing the findings of the previous study and confirming the accuracy of our predictions.
Promoters were categorized according to the expression characteristics of their parent genes. We identified 682 AAMφ-associated promoters, derived from genes up-regulated in WT-NeMφ relative to both WT-ThioMφ and IL4Rα−/−-NeMφ. The remaining 7135 promoters were classified as generic macrophage promoters. We compared AAMφ-associated promoters to the generic set, and identified only 3 over-represented TFBS motifs: MZF1, ZNF354c, and PPARG:RXRA (also known as PPAR response element [PPRE]; Table 2; Figure 6E). We used the Regulatory Sequence Analysis Toolkit36 to count the number of instances of each motif within our set of 682 AAMφ-associated promoters and found that 113 (17%) contained high-confidence PPRE elements. Because the MZF1_1-4 and ZNF354c consensus motifs are very short, we could not quantify these at the default significance threshold (P < .00001). The observation of an abundant and over-represented PPRE identifies PPAR-mediated transcription as a significant component of AAMφ-associated transcriptional regulation in vivo.
The 3 statistically over-represented position weight matricies in AAMφ-associated promoters relative to all macrophage promoters
| Jaspar motif . | Clover P . |
|---|---|
| MZF1_1-4 zinc-coordinating | .000 |
| PPARG::RXRA zinc-coordinating | .004 |
| ZNF354C zinc-coordinating | .006 |
| Jaspar motif . | Clover P . |
|---|---|
| MZF1_1-4 zinc-coordinating | .000 |
| PPARG::RXRA zinc-coordinating | .004 |
| ZNF354C zinc-coordinating | .006 |
Clover P value was determined by permutation test with 1000 iterations.
Analysis and quantification of NeMφ-derived eicosanoids
GO and KEGG GSEA identified augmented mitochondrial and metabolic gene expression, specifically the TCA cycle, as infection and IL4Rα-dependent processes (Figure 6A-B). Our analysis of AAMφ-associated cis-regulatory features further supported a role for PPAR involvement (Figure 6E). Together, these strongly suggest that PPAR-mediated transcription is a driving component of the AAMφ phenotype in vivo. PPARγ and oxidative phosphorylation are described as necessary for alternative activation,4 yet NeMφ PPARγ expression was low (Figure 6C). As PPARδ potentiates alternative kupffer cell37 and adipose tissue macrophage activation38 we hypothesized that it may compensate for PPARγ in NeMφ. The PPAR family are ligand-dependent transcription factors. Thus, although PPARδ was not DE, we explored the possibility that increased ligand availability might modulate PPARδ-dependent transcription in NeMφ.
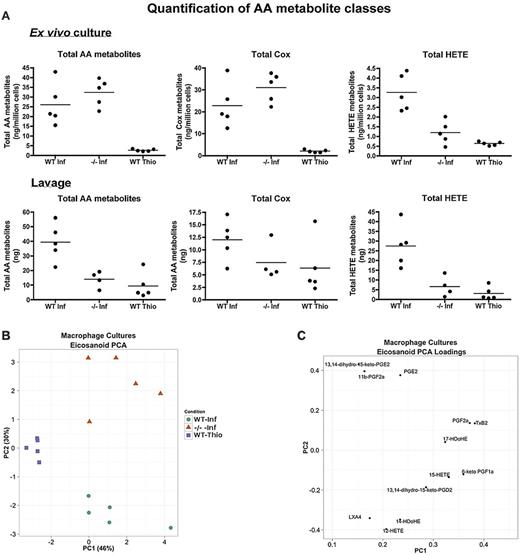
KEGG pathway analysis identified arachidonic acid (AA) metabolism, a known source of PPAR ligands, as up-regulated in WT-NeMφ (supplemental Table 6). Products of the AA enzyme cascade, eicosanoids, compose a wide variety of proinflammatory and anti-inflammatory mediators, including the prostaglandins, leukotrienes, and lipoxins. We characterized a broad range of these compounds produced by NeMφ and ThioMφ using LC-MS/MS in an independent, homologous experiment (lacking the IL4Rα−/− thioglycollate group). Individual eicosanoids (for a full list of species, see supplemental Methods) were measured in peritoneal lavage fluid and ex vivo in 12-hour cultures of adherence-purified macrophages. Ex vivo cultures showed that NeMφ produced significantly more eicosanoids than ThioMφ (Figure 7A) irrespective of IL4Rα expression. This cannot be attributed to enhanced activity of the AA-liberating phospholipase A2 enzymes as Pla2 transcripts were expressed in ThioMφ at a comparable level to NeMφ (supplemental Figure 12). This suggests that IL4Rα-independent effects of B malayi drive AA catabolism.
Characterization of AAMφ-derived eicosanoids. (A) Quantification of arachidonic acid (AA) metabolites in 12-hour cultures of purified macrophages (top) and in peritoneal lavages (bottom). (B) PCA scores plot for 12-hour cultures of purified macrophages. (C) PCA loading plot for panel B, showing how individual eicosanoids contribute toward the primary and secondary principal (x and y, respectively) axes. (D) Pie chart showing the breakdown in the production of cyclo-oxygenase metabolites between WT-NeMφ and IL4Rα−/−-NeMφ. (E) Schematic of the AA cascade reproduced using VANTED. Square boxes with red text represent genes; and rounded boxes with black text, measured metabolites. Black arrows indicate enzymatic reactions; and red arrows, auto-oxidation. Relative expression values (normalized such that the total over all 3 conditions = 1) are beneath the associated gene/metabolite. WT B malayi (left box) IL4Rα−/−B malayi implanted (center), and WT thioglycollate (right). (F-G) PCA scores and loading plot as in panels B and C for the peritoneal lavages.
Characterization of AAMφ-derived eicosanoids. (A) Quantification of arachidonic acid (AA) metabolites in 12-hour cultures of purified macrophages (top) and in peritoneal lavages (bottom). (B) PCA scores plot for 12-hour cultures of purified macrophages. (C) PCA loading plot for panel B, showing how individual eicosanoids contribute toward the primary and secondary principal (x and y, respectively) axes. (D) Pie chart showing the breakdown in the production of cyclo-oxygenase metabolites between WT-NeMφ and IL4Rα−/−-NeMφ. (E) Schematic of the AA cascade reproduced using VANTED. Square boxes with red text represent genes; and rounded boxes with black text, measured metabolites. Black arrows indicate enzymatic reactions; and red arrows, auto-oxidation. Relative expression values (normalized such that the total over all 3 conditions = 1) are beneath the associated gene/metabolite. WT B malayi (left box) IL4Rα−/−B malayi implanted (center), and WT thioglycollate (right). (F-G) PCA scores and loading plot as in panels B and C for the peritoneal lavages.
Principal component analysis (PCA) of eicosanoid profiles from cultured macrophages showed that the 3 treatment groups formed distinct clusters (Figure 7B). IL4Rα−/−-NeMφ produced high levels of proinflammatory PGE2 and downstream metabolites that promote acute phase inflammation.39 WT-NeMφ were characterized by production of the 12/15-lipoxygenase–derived HETE metabolites, predominantly 12-HETE (Figure 7C). Interestingly, by far the most abundant Cox derivative in WT-NeMφ was 6-keto-PGF1α, the auto-oxidation product of the PPARδ agonist prostacyclin (PGI239 ; Figure 7D).
Integrating the lipidomic profiles and gene expression data allowed us to better understand the relationship between transcriptional profiles and metabolic phenotype. We plotted gene expression and metabolite data onto a pathway map of the enzymatic cascade, relating AA catabolism to metabolite production.41 Alterations in synthase gene expression showed high concordance with the relative abundances of daughter metabolites (Figure 7E), with one notable exception. Despite robust expression of Gpx1, Alox5, and Lta4h, we were unable to detect any 5-lipoxygenase derived metabolites, 5-HETE 5-OxoETE, LTB4, or 20-OH LTB4 (supplemental Figure 13). Thus peritoneal macrophages were primed to produce leukotrienes but did not. Substrate competition for AA or higher-level control of leukotriene biosynthesis may explain this observation. In summary, eicosanoid production by macrophages is stimulated in response to B malayi. Rather than affecting total eicosanoid production, IL4Rα-dependent signaling modulated the architecture of the downstream enzymatic cascade, leading to a more anti-inflammatory eicosanoid profile.
Heterotypic interactions contribute toward the eicosanoid profile in vivo. To discern the relative contributions of macrophage- and non–macrophage-derived eicosanoids, we measured AA products in peritoneal lavage from the mice in the same experiment (supplemental Figure 13). PCA plots (Figure 7F) showed no distinction between WT thioglycollate-injected and IL4Rα−/−-B malayi–implanted mice. The WT B malayi–implanted individuals, however, clustered distinctly, showing that the response to B malayi implantation generated a unique, IL4Rα-dependent, eicosanoid environment. This also implies that B malayi is not a major source of total peritoneal eicosanoids in this model. WT-implanted mice had increased HETE, PGD2, and TxB2 concentrations (Figure 7G), the latter 2 of which are eosinophil chemoattractants.42,43 It is unlikely that the PGD2 was macrophage-derived as low levels were produced in AAMφ ex vivo cultures. TxB2 concentrations correlated with, and may be explained by, the increase in macrophage numbers associated with infection in a WT environment (supplemental Figure 14A-B). We have therefore identified a distinct lipid effector profile in the response to helminth infection. Although the activity of 5-lipoxygenase did not contribute toward this profile, the actions of cyclooxygenase and 12/15-lipoxygenase did.
Discussion
We have characterized the phenotype of B malayi–elicited AAMφ using RNA-Seq and a targeted lipid analysis. Together, these reveal the effects of IL4Rα-dependent signaling on macrophage physiology in a chronic Th2 environment. Our findings strongly support the emerging paradigm that alternative activation and inflammation are associated with wholesale alterations in metabolism.4 Further, GSEA and a detailed analysis of AAMφ-associated chemokine and cytokine expression provide intriguing clues as to the physiologic role(s) of AAMφ in vivo.
One of the more unexpected findings of our analysis was that, collectively, macrophage complement genes were among the most abundantly expressed and differentially regulated in the response to helminth challenge and may represent a previously unrecognized effector axis of AAMφ. A requirement for complement in antihelminthic immunity has previously been demonstrated, as C3 mediates clearance during both Strongyloides stercorallis44 and Schistosoma mansoni infection.45 In the context of B malayi, complement interacts with microfilariae46 and may contribute toward their clearance. Here we identify AAMφ-derived Fcna as a candidate for further study into the regulation of the complement response to filarial nematode infection.
We also identified IL4Rα-dependent expression of cytokines not currently associated with AAMφ physiology, including Bmp6 and Wnt2. Intriguingly, Bmp6 has been associated with increased macrophage iNOS activity, suggesting that it may curtail alternative activation.47 Wnt2 is a highly abundant AAMφ-derived cytokine with no identified role in macrophage biology. Wnt2 does, however, affect hematopoietic lineage commitment,48 and our AAMφ-up clusters contained Cdh1 (E-cadherin), an AAMφ marker in both humans and mice.49 Given the known convergence between cadherin and Wnt pathways, further work to elucidate the role of Wnt2 in macrophage biology is warranted. We have observed IL4Rα and infection-dependent production of cytokines, chemokines, and eicosanoids, suggesting that NeMφ aid in the orchestration and maintenance of the immune environment. Specifically, NeMφ potentially regulate eosinophil recruitment via multiple, redundant, mechanisms. Reese et al previously demonstrated chitin-induced eosinophilia as an AAMφ-dependent process,50 but whether they fulfill this role in the more complex environment of live parasite infection is unclear. We show here that WT-NeMφ transcribe eosinophil chemotactic factors in vivo. Ccl816 and Ccl2417 operate as Ccr3 ligands, whereas TxA2,43 the bioactive precursor of TxB2, induces eosinophilia by affecting vascular epithelial integrin expression. Our data are therefore consistent with AAMφ-mediated recruitment and maintenance of eosinophilia during helminth infection. Interestingly, in contrast to the work by Reese et al,50 our proposed mechanism for eosinophil recruitment is LTB4-independent. Our findings may differ because Reese et al used an acute stimulus, whereas we use a chronic model of helminth infection. Nevertheless, our data suggest cooperative interactions between AAMφ-derived protein and lipid mediators ensure robust, tightly regulated eosinophil recruitment to sites of infection.
IL4Rα-dependent signaling led to reduced expression of multiple proinflammatory cytokines and chemokines, supporting a model wherein AAMφ are anti-inflammatory or noninflammatory. Macrophage PPAR activity augments alternative activation and reduces the expression of proinflammatory cytokines.4 Indeed, thiazolidenediones, antidiabetic PPARγ agonists, improve insulin sensitivity in obese white adipose tissue (WAT) partly by curtailing CAMφ.4 Global similarities can be drawn between our surveyed NeMφ and the beneficial AAMφ in WAT of lean individuals, including enhanced oxidative metabolism and low proinflammatory cytokine expression.4 By assaying TFBS in NeMφ promoters, we provide compelling evidence that PPAR-mediated transcription is an important facet of the macrophage response to helminth infection in vivo. WAT AAMφ and NeMφ are however distinct subsets. WAT AAMφ express PPARγ, Il10, and Cd36,51 whereas Cd36 and PPARγ were down-regulated, and Il10 was not expressed, in NeMφ (supplemental Table 3). This is consistent with alveolar AAMφ failing to express IL10 in response to Nippostrongylus brasiliensis.52 However, the observation that N brasiliensis–infected mice show improved glucose tolerance53 demonstrates that the physiologic consequences of modulating alternative activation, either in the response to infectious disease or during homeostasis, imparts similarities that are greater than the observed differences between WAT AAMφ and NeMφ. This is perhaps not surprising as alternative activation in both these disparate environments is STAT6-dependent. An improved understanding of the influence of context-specific (ie, tissue/infection model) cues in fine-tuning AAMφ polarization is required to fully appreciate the functional diversity of AAMφ phenotypes.
We observed low PPARγ expression in NeMφ, consistent with a recent report of PPARγ expression profiles in different macrophage subsets.54 Analysis of AAMφ-associated cis-regulatory regions suggested a role for PPARs in WT-NeMφ, and we therefore sought a mechanism for PPAR-dependent transcription during B malayi infection. By profiling macrophage-derived eicosanoids, we found that nematode-, but not thioglycollate-elicited, macrophages abundantly produced the PPARδ ligand PGI2. Because of the low expression of PPARγ in NeMφ and constitutive PPARδ expression, macrophage PGI2 production provides a feasible and testable explanation for the maintenance of PPAR-dependent transcription in NeMφ. However, it should be noted that, because NeMφ do express PPARγ, we cannot rule out a role for both these transcription factors during alternative activation. We have shown that macrophage eicosanoid generation is IL4Rα independent in this system. This suggests that the liberation of PPAR ligands is B malayi–mediated and is perhaps achieved via a pathogen associated molecular pattern-dependent mechanism. IL4Rα-dependent signaling did, however, alter the expression of AA catabolic enzymes, enhancing the production of anti-inflammatory HETEs.3 Hence, extracellular cues (ie, IL4/IL13) manipulated the architecture of the enzymatic cascade, reorganizing the eicosanoid landscape. Ultimately, this reduced proinflammatory eicosanoid production and increased the synthesis PGI2 and anti-inflammatory HETEs. We propose a model in which the presence of a “danger signal” is coupled with cytokine-dependent structural reorganization, providing both an initiator and higher-order specificity that contributes toward sustaining the NeMφ phenotype in vivo.
We have defined the phenotype of IL4Rα-stimulated AAMφ in vivo during the response to a nematode infection. All of the data we present are supportive of anti-inflammatory roles for AAMφ in this setting. To summarize, WT-NeMφ broadly down-regulated the expression of numerous proinflammatory cytokines and chemokines. The eicosanoid environment generated by NeMφ is defined by enhanced production of anti-inflammatory HETE metabolites and also the abundant production of the PPARδ ligand PGI2. We infer that NeMφ have a decreased migratory capacity because of the down-regulation of multiple chemokines and therefore are unlikely to prime lymphatic T-cell responses. Our study has also identified the production of complement as a major component of the macrophage response to filarial nematode challenge. Although complement may aid in the recognition and clearance of either adult B malayi or microfilarae, it also has extensive immunoregulatory functions.55 For example, complement C5a interacts with surface Fc receptors to modulate inflammation.55 In this context, our WT-NeMφ strongly up-regulated expression of the regulatory Fcgr2b (supplemental Table 3). Thus, the complement-Fc receptor axis induced by AAMφ in the response to nematode challenge may be an additional anti-inflammatory feature of IL4Rα-dependent macrophage activation.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Marian Thomson in the GenePool Genomics facility for support in RNA-Seq data generation, Martin Waterfall for expertise with flow sorting, and Tom Freeman, David Hume, Stephen Jenkins, and Lucy Jones for critical reading of the manuscript.
This work was supported by the Wellcome Trust (PhD studentship award, G.D.T.) and the MRC (program grant MRC-UK G0600818, J.E.A.; and core funding to the GenePool facility MRC 60900740, M.L.B.). B.H.M. and P.D.W were supported by the European Regional Development Fund, Highlands and Islands Enterprise, and the Scottish Funding Council.
Wellcome Trust
Authorship
Contribution: G.D.T., D.R., M.L.B., and J.E.A. devised experiments; G.D.T. performed most of the experimental work and analyses; B.H.M., P.D.W., and G.D.T. performed lipidomics; G.D.T., M.L.B., and J.E.A. wrote the manuscript; and all authors contributed to writing the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Mark L. Blaxter, Institute of Evolutionary Biology, School of Biological Sciences, University of Edinburgh, Edinburgh, EH9 3JT, United Kingdom; e-mail: mark.blaxter@ed.ac.uk; and Judith E. Allen, Institute of Immunology and Infection Research, School of Biological Sciences, University of Edinburgh, Edinburgh, EH9 3JT, United Kingdom; e-mail: j.allen@ed.ac.uk.